1. Background
Azoospermia is one of the most serious reproductive disorders that occurs in 1% of the male population. Various factors are involved in azoospermia; however, its precise etiology is almost unknown (1). Most of these factors are complex gene-environment interactions (2). This reproductive-related disorder is classified as obstructive azoospermia (OA), induced by ejaculatory pathway obstruction, and non-obstructive azoospermia (NOA) caused by a failure in spermatogenesis (3). NOA is a disorder of genetic origin that is represented as a failure of spermatogenesis within the testis, including various reasons, such as chromosomal abnormalities and Y chromosome microdeletions (4). This phenotype has different causes (affecting various functions, such as differentiation of gonads, the activity of the hypothalamic–pituitary axis, and spermatogenesis), making it a complex and heterogeneous disorder (5). Essential genes involved in spermatogenesis are placed on the proximal area of the Y chromosome (Yp11). Azoospermia factor (AZF) is located in this region, which includes sub-regions of AZFa, AZFb, and AZFc (6). Ubiquitin-specific peptidase 9, Y-link (USP9Y), is a functional gene on the AZFa region with a DNA of 170 kb and at least 46 exons. USP9Y is responsible for encoding a protein (7) that acts as ubiquitin C-terminal hydrolase, which its elimination is related to azoospermia or violent oligospermia (8). USP9Y expression is limited to spermatids, and point mutations in this gene can result in various clinical phenotypes of male infertility, including spermatid maturation arrest, oligospermia, or asthenozoospermia, linked to decreased sperm cell mobility and concentration (9). Also, complete loss of the gene is associated with NOA (10, 11). In many complex diseases, genetic susceptibilities have been identified through genome-wide association studies (GWAS) using single-nucleotide polymorphism (SNP) arrays (12, 13). There is not enough information about the relationship between the USP9Y gene polymorphisms and azoospermia; thus, this study aimed at detecting the relationship between 5 polymorphisms of USP9Y and azoospermia susceptibility in the target population.
2. Objectives
Because different genes are responsible for the process of spermatogenesis and the formation of azoospermia, it is important to evaluate the genetic basis of infertility. In this study, we focused on the probable communication between the polymorphisms in the USP9Y gene and NOA.
3. Methods
This case-control study was approved by the Academic Center for Education, Culture, and Research (ACECR), Qom province, and the Biomedical Research Ethics Committee (IR.ACECR.JDM.REC.1397.015).
Regarding the azoospermia frequency in the population (1%), and the earlier findings, the minimum sample size was calculated based on the following formula (14):
Accordingly, 200 men attending the infertility treatment center from August 2016 to August 2017 were divided into the case (NOA) and control groups, and written informed consent was obtained before enrolling in the study. The inclusion criteria were as follows: Participants aged 25 - 35 years old and being physically and anatomically healthy with normal karyotype (Table 1). Healthy people for the control group had at least one healthy child with no more than two years passing since the birth of the last child. Semen from both cases and controls were analyzed according to WHO protocol, and urological examinations were performed for anatomical integrity of the genital system. Also, the NOA of the cases was approved by an urologist. Patients with anatomic disorders of genitalia, testis neoplasms, chromosomal numerical and structural abnormalities, or Y chromosome microdeletions were excluded.
| Karyotype | 46, XY |
|---|---|
| Microdeletion chromosome Y | NO |
| Spermogram with centrifuge | No sperm |
| TESE | No sperm |
| Age (y) | 25 - 35 |
| Varicocele | No |
| Gonadotropins (chemotherapy/radiation) | No |
Inclusion Criteria for Non-obstructive Azoospermic Men
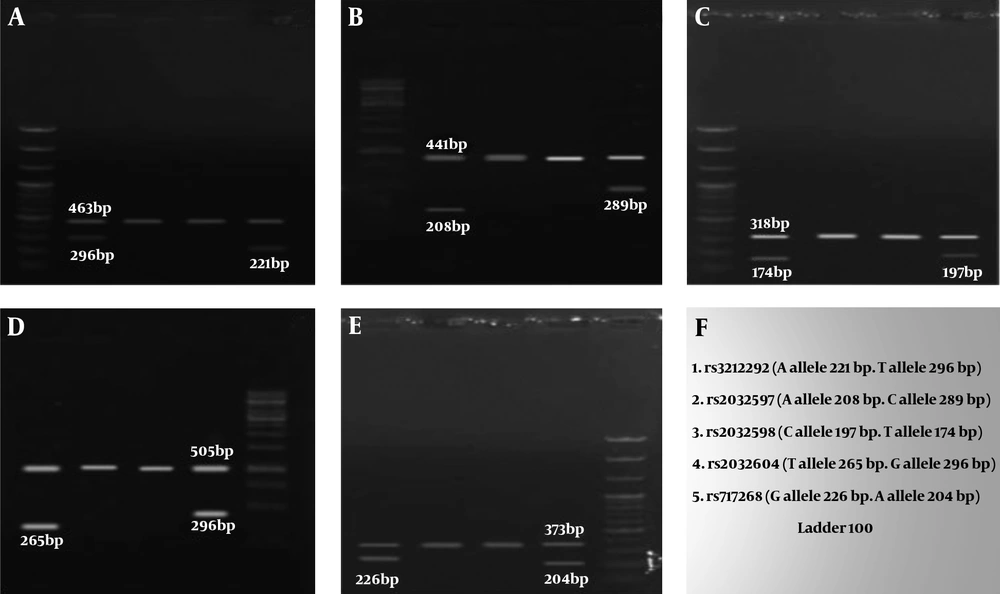
In order to confirm NOA, testicular biopsies of patients who had no sperm according to their semen analysis were assessed by a pathologist. Furthermore, serum levels of LH, FSH, and testosterone were measured using an ELISA kit in the case and control groups. Genomic DNA was extracted from blood samples using GeneAll® ExgeneTM kit. We used the tetra-primer amplification refractory mutation system- polymerase chain (PCR) method, in which two pairs of primers are used to analyze USP9Y rs2032597 (A>C), rs2032604 (T>G), rs2032598 (T>C), rs717268 (G>A), and rs3212292 (T>A) genotypes. Primers were designed by Primer1 online software (http://primer1.soton.ac.uk /primer1 .html). PCR primers are given in Table 2.
| Rs | Nucleotide Sequence | PCR Product (bp) |
|---|---|---|
| rs3212292 | FO-RO = 463; FO-RT = 296; RO-FA = 221 | |
| FA (A allele) | 339 GCTTGGGATTTTTCTCCTGGACATCA 364 | |
| RT (T allele) | 391 ACCTTAAAGCAATCAAAAAGATGATGAA 364 | |
| FO (5' - 3') | 96 TGCCAGGTATTAAATGACAGTTCTAAAGCA 125 | |
| RO (5' - 3') | 558 TGATTGGAAGAAACAACAATGACCAATT 531 | |
| rs2032597 | FO-RO = 441; FO-RC = 289; RO-FA = 208 | |
| FA (A allele) | 496 TTTGTTAAATAATTTCATGTTTGTCCA 522 | |
| RC (C allele) | 550 ACATAACTTAATAAGAGCTGCAATTACTG 522 | |
| Forward outer primer (5' - 3') | 262 TAAATTTTCCATTTCTAGTATGCTTCAC 289 | |
| Reverse outer primer (5' - 3') | 702 AAAAGAACTAAACTTGCCAATTACTTTC 675 | |
| rs2032598 | FO-RO = 318; FO-RT = 174; RO-FC = 197 | |
| FC (C allele) | 331 ATAATGGCCAGCAATTTAGTATTGACC 357 | |
| RT (T allele) | 382 AACAGCACATGCATTAGTAAAAGGCA 357 | |
| Forward outer primer (5' - 3') | 209 AAAGAAAAGGCTCTTACATTACAGGACC 236 | |
| Reverse outer primer (5' - 3') | 526 GAGCAAGATTCCATCTAAAAACAAAACA 499 | |
| rs2032604 | FO-RO = 505; FO-RG = 265; RO-FT = 296 | |
| FT (T allele) | 493 TATCCCCCAAACCCATTTTGATGCCTT 519 | |
| RG (G allele) | 547 GAAAATAATAATTGAAGACCTTTTAAATC 519 | |
| Forward outer primer (5' - 3') | 252 AAGATGTTAAAGAGGCCAGCTTTGTG 277 | |
| Reverse outer primer (5' - 3') | 756 GGTTACAAAGAACATATCACAATGGCAA 729 | |
| rs717268 | FO-RO = 373; FO-RA = 204; RO-FG = 226 | |
| FG (G allele) | 234 ATAGGATGTATTATTTTCTATGATTCTGAG 263 | |
| RA (A allele) | 289 TGTTCCTTTACATATGAGATGTATGTT 263 | |
| Forward outer primer (5' - 3') | 86 AACTTCTGAGTGAATTTCTTAGTTGTAG 113 | |
| Reverse outer primer (5' - 3') | 458 CATAGTACTCAGATGCTGATTATACAAA 431 |
Nucleotide Sequences for Designed Primers
Statistical analysis for genotype and allele frequencies, azoospermia association, and hormonal evaluation were performed by the χ2 and paired t-test using SPSS software. A P-value of less than 0.05 was considered to be statistically significant.
4. Results
The mean age of the control and case subjects was 31.12 ± 1.38 and 31 ± 1.688 years, respectively. Table 3 presents the frequencies of genotypes and alleles in 5 SNPs. The χ2 test showed that there was no significant difference between the genotypic and allelic frequencies between the case and control groups, and these genotypes did not increase the chance of disease or showed no protective effect against NOA. The electrophoresis results of 5 SNPs are shown in Figure 1.
| rs/Genotype & Allele | Non-obstructive Azoospermia, No. (%) | Control, No. (%) | P-Value | OR (95% CI) |
|---|---|---|---|---|
| rs3212292 (T>A) | 0.346 | 1.568 (0.612 - 4.019) | ||
| A | 12 (12) | 8 (8) | ||
| T | 88 (88) | 92 (92) | ||
| rs2032597 (A>C) | 0.744 | 0.899 (0.474 - 1.705) | ||
| A | 76 (76) | 74 (74) | ||
| C | 24 (24) | 26 (26) | ||
| rs2032598 (T>C) | 0.313 | 1.405 (0.724 - 2.728) | ||
| C | 26 (26) | 20 (20) | ||
| T | 74 (74) | 80 (80) | ||
| rs2032604 (T>G) | 0.192 | 1.784 (0.742 - 4.292) | ||
| G | 15 (15) | 9 (9) | ||
| T | 85 (85) | 91 (91) | ||
| rs717268 (G>A) | 0.39 | 1.652 (0.521 - 5.236) | ||
| A | 8 (8) | 5 (5) | ||
| G | 92 (92) | 95 (95) |
Allele and Genotype Frequencies of rs508485 (T>C) Its Association with the Non-obstructive Azoospermia Risk
The mean FSH level in the control and case group was 7.43 ± 1.947 and 16.88 ± 2.188 mIU/mL, respectively. The Mann-Whitney U test showed that the observed mean difference was significant (P < 0.001), and the FSH hormone increased in cases. The mean LH level was 5.7 ± 1.828 and 12.18 ± 1.471 mIU/ml in the control and case groups, respectively, and the observed difference between the two groups was significant (P = 0.002). The mean testosterone level in the control and case group was 4.6 ± 1.262 and 4.83 ± 1.292 ng/dL, respectively. The independent t-test indicated that the difference in mean testosterone level observed between the two groups was not significant (P = 0.849) (Table 4).
| Hormone | Control | Non-obstructive Azoospermia | P-Value |
|---|---|---|---|
| FSH | 7.43 ± 1.95 | 16.88 ± 2.19 | < 0.0001 |
| LH | 5.7 ± 1.82 | 12.18 ± 1.47 | 0.002 |
| Testosterone | 4.6 ± 1.26 | 4.83 ± 1.29 | 0.849 |
Comparison of Hormonal Levels in the Control and Non-obstructive Azoospermic Men
5. Discussion
Infertility is a multifactorial issue in which various factors, such as the environment and genetics, can be influential. Studies have shown that 50% of infertilities are related to men. The most important factors in male infertility are genetic factors that affect important physiological processes, such as hormonal homeostasis, spermatogenesis, and sperm quality (15). About 200 genes have been found so far that can regulate spermatogenesis, 30 of which are on the Y chromosome. Two major genetic factors in men's infertilities are Y chromosome microdeletions and chromosomal abnormalities (16, 17). Around 25% of NOA cases can be explained by single genetic abnormalities (including chromosome aberrations and point mutations), with the remaining patients considered idiopathic. It has been indicated that idiopathic NOA has a multifactorial etiology, in which both environmental and genetic factors may contribute to the development of the disease. In these cases, wide variations in the human genome, mainly single nucleotide polymorphisms (SNPs) and copy number variants (CNVs), are likely to confer genetic predisposition, complicating the elucidation of the underlying pathological mechanisms (18, 19). According to the influential advances in infertility treatment using assisted reproductive techniques (ART), which increase the likelihood of transferring genetic abnormalities to the next generations, identification of genetic abnormalities sounds valuable (20). In fact, the frequency of male infertility varies among different geographical regions and ethnic groups. It was found that among azoospermia cases, 83.7% were due to NOA (21). Also, Fogle et al. showed that 93% of azoospermia cases were recorded as NOA (22). However, the most geographical and ethnic reports in male infertility are on Y chromosome microdeletions.
Malekasgar and Masoudi showed that 51.6% of azoospermic infertile men and 52.6% of severe oligozoospermic patients from north Iran carry Y chromosome microdeletions (23, 24). In the Iranian population, the Y microdeletion frequency, according to the databases from 2003 - 2012, has shown variable ranges from 5 - 52%, which is within the worldwide range (15). This wide range of variability may be the consequence of different factors, such as the sample size, type of patient selection, and the variances among the populations (25). Also, Omrani et al. reported 24% microdeletions in infertile men from West Azarbayjan (26). It should be noticed that our study was performed on 200 participants referring to the infertility treatment center of ACECR in Qom province. One of the genes involved in the spermatogenesis process is the USP9Y gene, which was selected to examine in this study. Deletions affecting USP9Y were initially highly associated with mild to severe azoospermia or oligozoospermia (27). It was later documented that partial deletion of USP9Y in men is correlated with the milder infertility phenotype, indicating a minor effect for USP9Y in spermatogenesis (28). A study by Sun et al. on 576 infertile men and 96 healthy men showed that single-gene deletion associated with spermatogenic insufficiency occurred in USP9Y (29). Thus, USP9Y is not required for normal spermatogenesis, but its effects on fertility may be effective in combination with other genes in the AZFa region (30). In 2000, Foresta et al. analyzed 173 infertile men focusing on the AZFa region and found deletions in 9 patients, of whom one case showed deletions in the USP9Y gene (31). There is contradictory evidence about the impact of this gene on infertility and spermatogenesis, and the association between polymorphisms in the USP9Y gene and azoospermia has also not been investigated. We tested 5 polymorphisms of the USP9Y gene in this study. Some SNPs of this gene have been evaluated and shown to be associated with some other diseases. In a study, Wang and Park showed that the risk of schizophrenia, autism, and prostate cancer could be increased by rs2032598 and rs2032604 SNPs; however, no population-based study on azoospermia has yet been performed (32, 33).
It is of particular interest that the ubiquitin protein encoded by the USP9Y gene plays an important role in infertility and can affect the polymorphism of the USP9Y gene in male infertilities (34, 35). Studies have shown that the levels of sex hormones, such as LH, FSH, and testosterone were significantly higher in infertile men in comparison with healthy men. These hormonal changes were much more seen in azoospermic men than in other infertile subgroups (36, 37). These results suggest that the FSH, LH, prolactin, and testosterone levels cannot predict microdeletions in AZF in infertile men. More studies with a larger sample size should be considered to assess these microdeletions (38). Zhao et al. stated that sperm density, total sperm count, number of motile sperm, and percentage of motile sperm were positively associated with the testicular volume (39). However, our results in this study showed a decrease in FSH and LH hormones in the case group compared with the control group. Also, testosterone hormone levels in the case and control groups did not show a significant difference.
5.1. Conclusion
In general, our results showed that in the studied population, the frequency of genotypes and alleles did not play a role in developing NOA. The results of this study showed that one of the important reasons for NOA can be hormonal disorders so that in NOA, the levels of FSH and LH hormones are increased, which indicates that in people with sperm disorders, hormone levels should be checked at first, and then other tests and more detailed investigations should be regarded.