1. Background
Microsporidia species, as an obligatory intracellular protozoan, can infectall unicellular organisms of arthropods and fish, as the most common hosts of the parasite (1, 2). Microsporidia can penetrate the cytoplasmic myofibrils of host cells, which causes the destruction of muscular tissue and replace connective tissues in these areas. So, the affected tissue would have an emaciated and concave appearance (3, 4). Moreover, growth deformities result in decreased productivity, and even death (3, 5, 6). Also, this opportunistic parasite may cause the freezer-burn appearance of meat, which makes it inedible for humans (4), and economic losses in the aquaculture industry (1). This spore-forming parasite affects the host tissues, and form a cyst that may be restricted by the connective tissue of the host (4). More than 100 species of Microsporidia belonging to 21 genera of fish microsporidians have been identified (1, 2). Dasyatispora, Heterosporis, Pleistophora, Kabatana, and Microsporidium (2, 7) are the most widely identified species in fish. However, microsporidian infections are not limited to aquatic animals. So that, infections in the gastrointestinal tract (8) and muscle (9) of both immunocompromised and healthy individuals are reported (10). Traditionally, Microsporidian species are classified into groups or clades based on the morphological, structural differences, habitat, and host species (11). With the advancement of molecular genetics techniques, recently conducted phylogenetical studies have challenged the taxonomic classification of Microsporidia spp (11-13). In a way, the findings regarding phylogenetic and ultrastructural characteristics are sometimes contradicted with the morphological features of the parasite (14). Hence, using a combination of the host type, ecology, pathology, ultrastructural morphology, and phylogenetic methods to describe the novel microsporidia is recommended (11, 12). Studies conducted to classify unknown species of microsporidia have frequently used ribosomal RNA genes (3, 4, 13-15). Today, the small subunit ribosomal RNA (ssrRNA)-based phylogenetics is used as the main distinctive criterion among microsporidia species (Stentiford et al., 2013), and the taxonomic classification is more investigated based on the sequence data of this region of the Microsporidia genome (14).
Lizardfishes that belong to the Synodontidae family have a cylindrical body shape and inhabit benthic biotopes (16). The microsporidian Pleistophora oolyticus (17), H. saurida (3), and Glugea sp (18) have been reported in lizardfish living in the Egyptian coast of the Red Sea (S. tumbil), Arabian Sea and the Persian Gulf, respectively. Few studies have been performed in Iran on microsporidians infecting aquatic hosts (18). According to the recent high microsporidians infection reported in different native species of fish, comperhensive studies are needed to support the identified morphological findings.
2. Objectives
This study intended to report a new microsporidium from Saurida undosquamis of Persian Gulf, Heterosporis sp. through morphological and molecular data beside of the phylogenetic aspects relationships to other fish microsporidian species.
3. Methods
3.1. Sampling and Microscopy
Fifty specimens of S.undosquamis (Synodontidae, Harpadontinae) (19) were bought from several local markets of Ahvaz County, southwest of Iran within 6 months. All fresh dead fish were directly transferred to the fish health laboratory of the Veterinary Faculty in a cold container. The abdominal cavity of fish was observed for the irregular whitish cyst-like. Grossly visible cysts were isolated and collected from the infected fish. This study was performed in accordance with Helsinki's ethical guidelines.
3.2. Gross and Light Microscopic Study
The structural characteristics of the collected cysts were observed grossly. Then a wet smear of samples from the visceral cavity was prepared. After staining of wet smear with Giemsa, (1:10 dilution for 20 min) according to the guideline’s dissection (20), Histopathologic sections (with 4 µm thickness) were provided by fixation of the cysts in 10% buffered formalin to study the structure of the cysts. Then, the arrangement of the spores within the cysts was investigated using a light microscope.
3.3. Electron Microscopy
Some small isolated cysts were fixed in 3% glutaraldehyde in phosphate buffer. The cysts were postfixed in 1% osmium tetroxide for two hours and washed with phosphate buffer. They were dehydrated through a graded acetone series and infiltrated in different resin in acetone concentrations of 50%, 67%, and finally pure resin. Ultramicrotomy (No. OMU3, C. Reichert, Austria) was performed on ultrathin sections of 60 - 80 nm thickness. Then specimens were mounted on copper grids and stained with uranyl acetate and lead citrate and investigated under the transmission electron microscopy (TEM) (Leo model 909) (21). Some cysts were also prepared for scanning electron microscopic (SEM) study (SC7620 sputter coater - Leo 1455 VP SEM, Germany) by usual methods.
3.4. Genomic DNA Extraction; PCR Amplification and Sequencing
Cysts were collected from fish and washed several times with double-distilled water. These cysts containing spores were dissected by scalpel, homogenized and the genomic DNA was extracted using the DNPTM kit (SinaClon, Tehran, Iran) according to the manufacturer's instructions. The extracted DNA was kept frozen at -20 for future use. The amplification of the most part of the small subunit ribosomal RNA gene was performed by PCR using the primers V1f (5’- CACCAGGTTGATTCTGCC-3’) and 1942r (5’-GGTTACCTTGTTACGACTT-3’) (22). The PCR reaction was amplified in a BioER Life TouchTM PCR Thermal Cycler (BIOER, Hangzhou Bioer Technology Co., Ltd, Japan) under initial denaturation at 94°C for 7 min, followed by 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 2 min. The final elongation step was performed at 72°C for 10 min. The PCR products were electrophoresed through a 1% (agarose gel stained with safe stain (CinnaGen Inc, Iran), and results were visualized ultraviolet transilluminator. A 1kb DNA ladder (CinnaGen Inc., Iran) was run to estimate the size of DNA in each gel. The amplified SSU rRNA region was purified using the AccuPrep PCR Purification Kit (Bioneer, Daejeon, Korea) and sequenced by Bioneer Biotechnology company (Seoul, South Korea) in both directions. After considering the sequences, the data were analyzed using the ClustalW accessory application within BioEdit V. 7.0.5.3 software. The similarity of the microsporidian species was determined with other sequences in GenBank by the basic local alignment search tool (BLAST) program in NCBI databases (National Center for Biotechnology Information). Then 15 similar Marinosporidia rDNA sequences were selected for molecular and phylogenetic inferences from GenBank. Brachiolaalgerae (AY963290) was presented as the outgroup taxon.
3.5. Phylogenetic Analysis
The alignment sequences were analyzed by the Molecular Evolutionary Genetics Analysis (MEGA: X) software (23), using the default parameters for the molecular phylogenetic study of the species. The evolutionary history was inferred by using the Maximum Likelihood method and Kimura 2-parameter model (24). The tree with the highest log likelihood (- 4102.22) is drawn. Support values for internal nodes were estimated by a bootstrap resampling procedure with 1000 replicates.
4. Results
4.1. Gross and Light Microscopic Examination
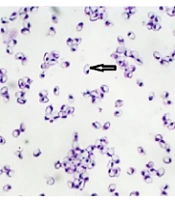
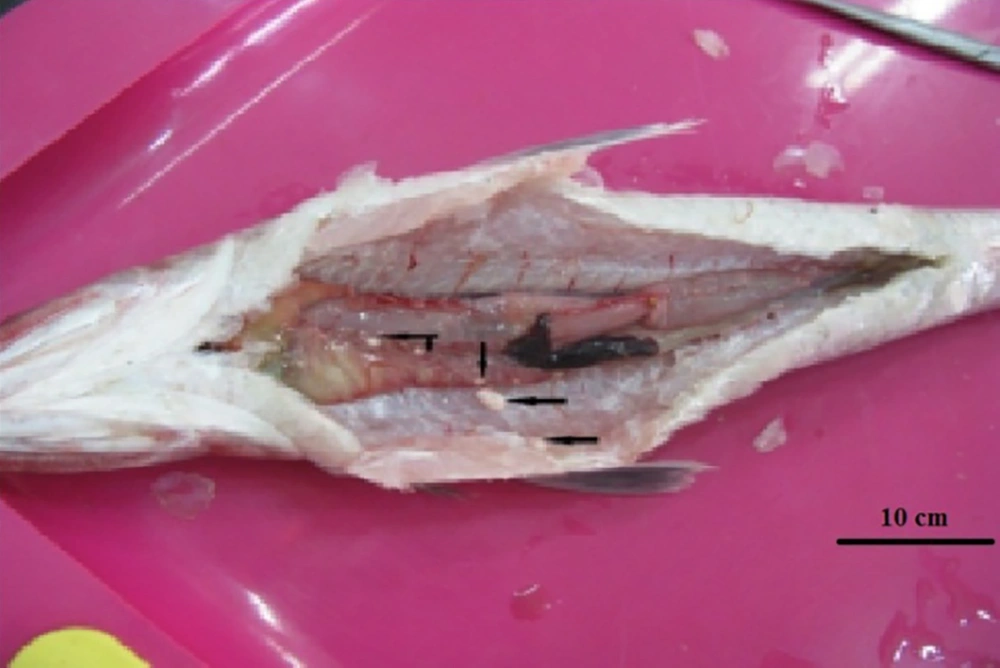
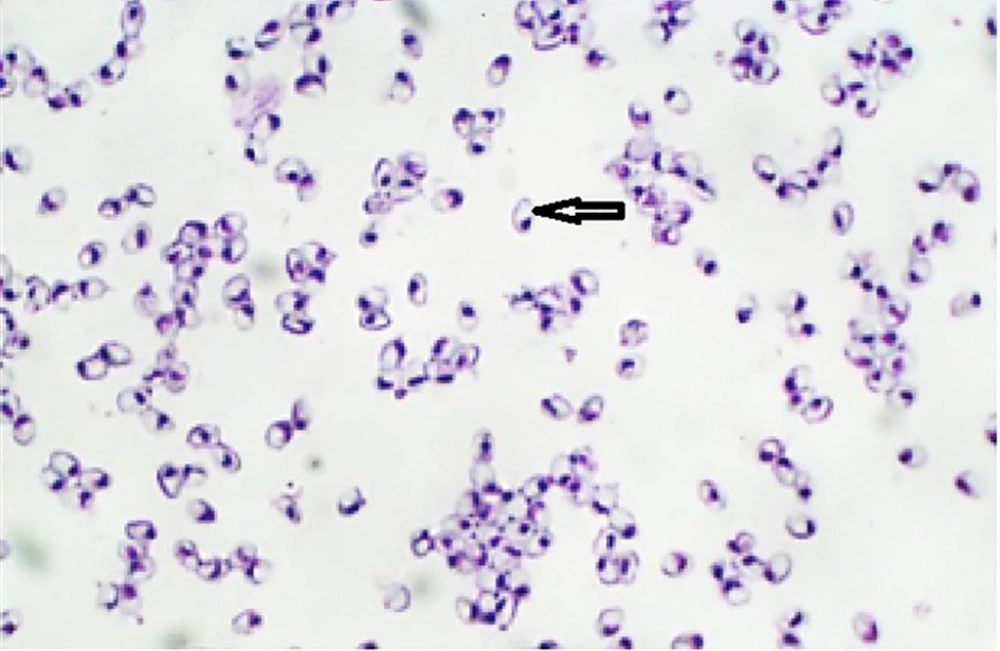
Twenty-two out of 50 of the examined fish were naturally infected with microsporidian parasites with an infected rate of 44%. Infection was confirmed as numerous macroscopic white cysts throughout the abdominal cavity, mesenteric tissues, and infrequently in the skeletal muscles or visceral organs of infected lizardfish (Figure 1). In wet smears, the spores were monomorphic and mostly ovoid with a large vacuole at the posterior end (Figure 2). Histological findings show that the large xenoma is encapsulated by a host-derived thick connective tissue due to the proliferation of microsporidian parasites.
4.2. Electron Microscope Observations
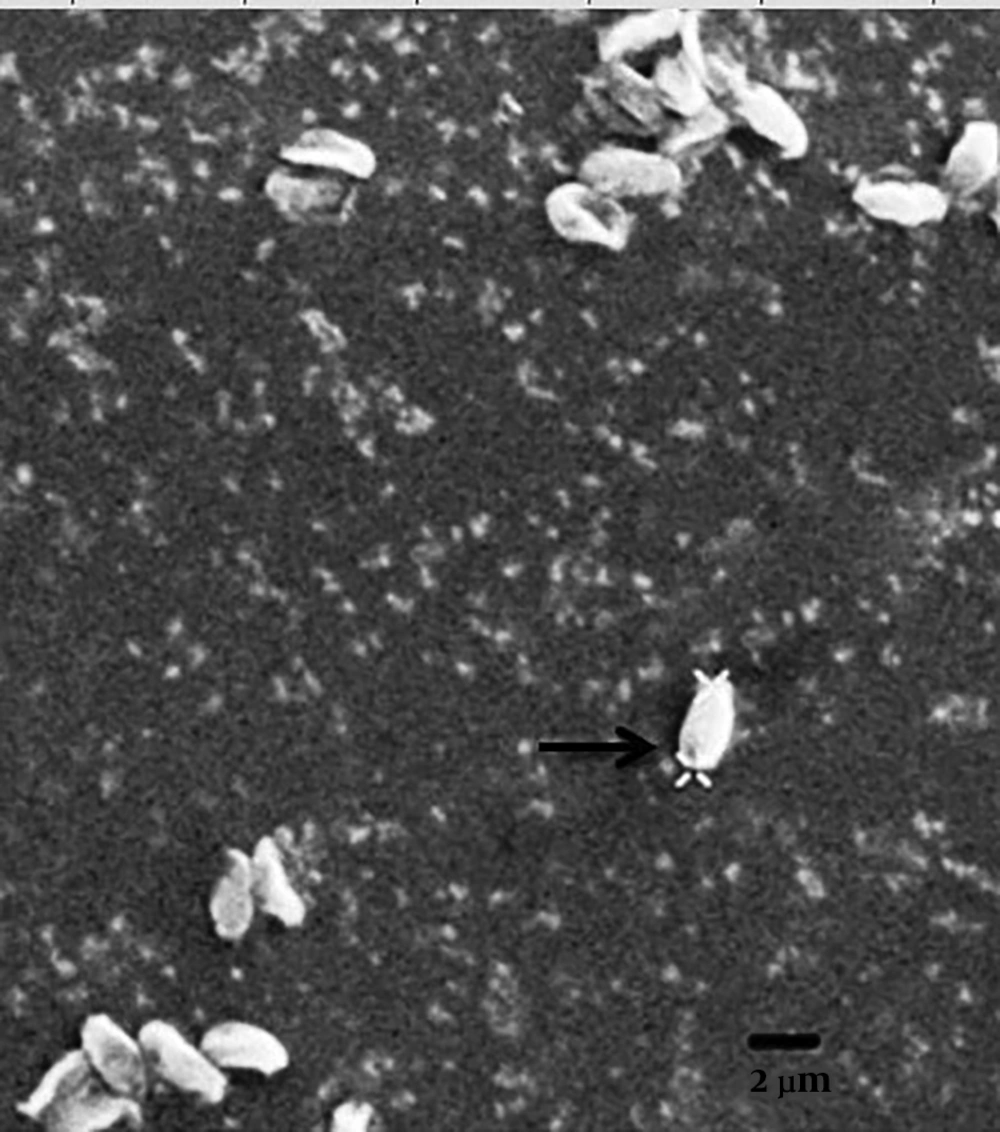
This parasite forms a xenoma structure. Unfortunately, we could observe only the mature microsporidian spores. The mature spores were monomorphic, uninucleated, and ovoid with the size of dime 2.8 - 2.9 μm long by 1.3 - 1.8 μm wide. The posterior vacuole was seen at the posterior end of most spores surrounding by 6-8 turns of polar filaments. The isofilar polar filament was located in one row with round-oval in cross-section. The spore wall was seen thick and amorphous and contained two layers of exospore and endospore. The anchoring disk was located in a central position at the anterior end of the spore and contacted the shaft of the future polar tube (Figure 3). They form sporophores vesicles (SPV) that surround sporoblast mature spores but not sporophorocyst’s (SPC) with a dense, solid wall surrounding all the developmental stages of the parasite.
4.3. Molecular Analysis
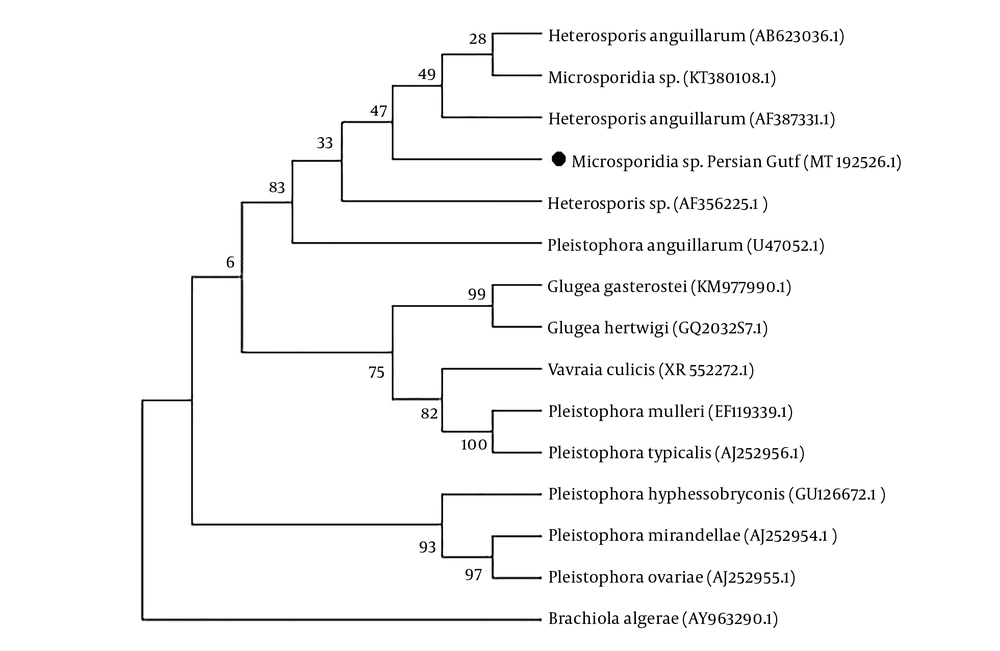
A 1,279 bp genomic sequence was amplified from the DNA of spores of the cysts and contained a partial sequence from the small subunit of ribosomal DNA (SSU rDNA) with accession number MT192526 in the GenBank database. Multiple sequence alignment revealed this lineage of fish-infecting microsporidia with high identities and BLAST scores belonging to the Heterosporis (Pleistophora) genera (bootstrap 83%). The maximum identity of the present sequence was shown as 99.05% (AF387331) (15), and 98.97% (AB623036) to H. anguillarum in nerve cells of Japanese eels (Anguillajaponica) (25), and also 99.19% (KT380108) to Microsporidia sp. Heterosporis-like in muscle cells isolated from Australian sea snakes (Hydrophiinae) (14).
Persian Gulf microsporidian is placed in a separate branch within a clade containing other species of Heterosporis. This clade is represented by Heterosporis spp. infecting predominantly Japanese eels (Anguillajaponica) from Australia (KT380108), Taiwan (AF387331) and (U47052), and Greater amberjack (Serioladumerili) from Japan (AB623036), and Yellow perch (Percaflavescens) from Finland (AF356225) (Figure 4).
5. Discussion
In the present study, the infection rate was high (44%), which is comparable to the other studies with an infection rate of 32.1% for H. saurida from S. undosquamis in the Arabian Gulf (3), 29.23% and 6.15% for H. lessepsianus from S. lessepsianus in winter and summer months, respectively, in the Red Sea (Egypt) (26), and also an infection rate of 30.6% for Pleistophorapagri from Pagruspagrus in the Red Sea, Egypt (27). These reports showed that marine microsporidiosis follows a temperature and seasonal pattern and the infection rate of disease increases during the cold months of the year. In this study, sampling was referred to the winter and spring months. As during the fish hibernation in cold months, fatigue and exhaustion dominate the fish, which makes them more susceptible to high degrees of parasitic infections (28). However, the pathogenesis role of each parasitic species should not be ignored. Also, the species of Pleistophora oolyticus (17), Glugea sp. (18), Microsporidium sp. (29), and Pleistophoraaegyptiaca (28), have been detected from different species of Saurida in the Egyptian coast of the Red Sea, the Persian Gulf of Iran, Saudi Arabia, and the Red Sea, Egypt, respectively. Today the classification of microsporidia species based on some ultrastructural features is not sufficient for diagnosing the microsporidian alone as well as the comprehensive application of molecular investigations. The Persian Gulf microsporidia sp. like G. anomola (20, 30), and G. vincentiae (31), formed xenoma structure surrounding by a thick wall of host connective tissue and the SPV without the formation of SPC but in some morphological features were different from Glugea species such as the shape and size of spore, and the number and arrangement of polar filament. The closest genetic species to the Persian Gulf microsporidian was H. anguillarum. Morphologically, it has been reported that H. anguillarum has SPV and no SPC but do not form xenoma structure while the Persian Gulf microsporidian had xenoma structure.
Here, the genomic study of the SSU rDNA region revealed similarities to species that was contradictory with the isolated species in structural characteristics. Miwa et al. reported H. anguillarum in farmed greater amberjack, Seriola dumerili, differs in some morphological characters such as observing no sporophorous vesicle, xenoma form of parasites, and having two different types of spores with our detected microsporidian (25). Also, Gillett et al. detected Heterosporis-like microsporidian from the sea snake that is morphologically inconsistent with isolated species (14). Although some structural characteristics of Persian Gulf microsporidian closely resembled Heterosporis species found in sea snake microsporidian, such as the presence of SPV and absence of SPC, but form monomorphic spores with different number and arrangement of polar tube coils and don’t have xenoma structure found in muscle tissue (14). Some species of Glugea and Heterosporis species spores can be in monomorphic, dimorphic, and even trimorphic size and shape ranges and with a different arrangement of polar tube coils (32), and have great phenotypic flexibility. The parasite plasticity and dimorphism would be scientifically acceptable in Marinosporidia class (12), that can be indicated the occurrence of the life cycle variation or multitrophic transmission among the class Marinosporidia within different hosts (14). Also, the similarity of SS rDNA of this species indicates a high rate of genetic conservatism in the house-keeping genes of microsporidians. Other gene sequences within this group may also need to be accurately investigated.
In this paper, the importance of a comprehensive survey including morphological and molecular features to detect microsporidian species has been shown.