1. Background
Canine morbillivirus or canine distemper virus (CDV) is responsible for canine distemper disease in many mammalian species and may cause systemic, respiratory, cutaneous, bony, or neurological symptoms in the affected animals (1, 2). CDV has been isolated from dogs and other Canidae members, including rodents, foxes, jackals, ferrets, coyotes, lions, tigers, rabbits, cheetahs, sea lions, and dolphins. Domestic dogs are the most common species infected with CDV. Although the disease has not been reported in domestic cats, experimental infection of specific pathogen free (SPF) cats has shown that the virus can multiply and cause lymphopenia without diffuse clinical symptoms (3-5).
CDV shows high lymphotropism, associated with the signaling lymphocytic activation molecule (SLAM) proteins in lymphocyte subsets. SLAM binding is required for lymphocyte infection, viral infection, and immunosuppression in CDV-infected patients (6, 7). It often leads to infections with opportunistic pathogens, such as Bordetella bronchiseptica, Candida spp., Clostridium piliforme, Toxoplasma gondii, Dirofilaria immitis, Mycoplasma cynos, and Talaromyces marneffei (8). Infection with CDV has been reported concurrently with other common viruses, such as canine parvovirus type 2 (CPV-2), canine alphaherpesvirus type 1, and canine adenovirus type 1 and 2 (CAdV-1 and CAdV-2). Recent reports have shown that CDV infection can lead to diseases caused by emerging viruses, such as canine kobuvirus, pneumovirus, and canine respiratory coronavirus (CRCoV). Genomic and molecular studies have shown the presence of canine pathogens in symptomatic and asymptomatic dogs (9-11).
One of the main causes of severe leukemia following infection with morbilliviruses is apoptosis in immune cells, followed by immunosuppression (12, 13). After the removal of the virus from peripheral blood, the reduced expression or supply of antigens and maturation of lymphocytes may result in the persistence of immunosuppression (4). CDV suppresses immunity by affecting CD4+ cells. Therefore, dogs with CDV are more sensitive to opportunistic pathogens. Also, other viral, bacterial, fungal, and parasitic infections are more likely to occur concurrently in these dogs (7, 14).
Bordetella bronchiseptica is a small, gram-negative, rod-shaped bacterium, which is responsible for infectious bronchitis in dogs and other animals (rarely in humans). This bacterium is highly similar to B. pertussis and can remain stable in the environment for a long time (15). It is known that B. bronchiseptica can affect variable hosts and cause serious diseases in dogs, pigs, and rabbits; it has been also reported in cats, horses, and waterfowl (16). Moreover, infection with B. bronchiseptica leads to the inadequate growth of turbinates and the tip of the nose. In dogs, the virus causes acute tracheobronchitis (9), usually characterized by dry cough. This bacterium, along with CAdV-2 or canine parainfluenza virus, can be the cause of kennel cough syndrome (16).
In rabbits, B. bronchiseptica is often found in the nasal cavity. Although this bacterium frequently causes asymptomatic infections, it is considered to be a predisposing factor for Pasteurella multocida infection; these two viruses can also infect the nasal passages concurrently (15). Cats infected with B. bronchiseptica usually have tracheobronchitis, conjunctivitis, rhinitis, mandibular lymphadenopathy, and pneumonia. However, rhinitis in cats may also be caused by Herpesviridae species, Caliciviridae species, Mycoplasma species, and Chlamydia psittaci (16).
2. Objectives
With this background in mind, the present study aimed to investigate concurrent infection with CDV and B. bronchiseptica, using genomic and immunological identification methods.
3. Methods
3.1. Sampling
Sampling was carried out among dogs, which were referred to Isfahan clinics between the spring of 2018 and the winter of 2019. The animals were chemically immobilized using a combination of ketamine (Alfasan, Netherlands) and acepromazine (Hoogstraten, Belgium). After recording their age and sex, a 3-mL blood sample was taken from the saphenous or cephalic vein and transferred to laboratory for serum isolation. The separation of serum and buffy coat from blood was performed by centrifugation at 3000 rpm for 15 minutes. Simultaneously, an eye sample, a respiratory swab, and a gastrointestinal tract swab were prepared from the immobilized animals and added to sterile phosphate-buffered saline (PBS). The samples were kept at -70°C until further tests. Overall, in this study, 50 buffy coat samples were taken from dogs with symptoms of respiratory and gastrointestinal tract involvement, and 50 samples were taken from seemingly healthy dogs.
3.2. Rapid Immunochromatography for CDV
Rapid immunochromatography was used for the initial diagnosis of CDV. Generally, eye discharge, saliva, urine, and blood samples can be used to diagnose distemper, using rapid immunochromatography kits. In the present study, eye samples and respiratory swabs were used to identify CDV by the VetALL SensPERT Canine Distemper Virus Test Kit (CDV Ag, New Zealand). Briefly, the swab was placed in a buffer prepared by the kit and shaken several times to dilute the sample. Next, using an emitter embedded in the kit, 100 µL of the test sample solution was drawn (approximately 4 drops) and was poured into an elliptical hole for loading. The time was recorded from this point on. After adsorption of the sample solution on the test plate pad, the emergence of two red bands at C (control) and T (test) positions indicated a positive test result.
3.2.1. Nucleic Acid Extraction
For the detection of CDV, RNA was extracted using a QIAamp Viral RNA Mini Kit (Cat. No.: 52904; Qiagen, Germany), based on the manufacturer’s instructions. For investigation of B. bronchiseptica, total DNA was isolated with some modifications in the protocol proposed by Doyle et al. (cited in Davies et al.) (17). Briefly, the samples were centrifuged, and the supernatants were discarded. Next, treatment with an extraction buffer was performed by incubating and shaking at 65°C for 30 minutes. The mixture was then subjected to chloroform-isoamyl alcohol (24:1), and after centrifugation, isopropanol was used to precipitate DNA. After centrifugation and elimination of isopropanol, washing was performed twice by adding 70 µL of 70% ethanol. Finally, the DNA pellet was dried at room temperature; re-suspended in 30 µL of deionized sterile water; and placed at 37°C for 30 minutes.
3.2.2. Reverse Transcription
The cDNA synthesis was performed using a TaqMan RT Kit (Cat. No.: N8080234; Invitrogen, USA). The reverse transcription (RT) Master Mix consisted of 1 µL of 10X RT buffer, 2.4 µL of 25 mM MgCl2, 1.8 µL of 10 mM dNTPs, 0.4 µL of random hexamers, 0.3 µL of RNase inhibitors, 0.3 µL of reverse transcriptase (50 U/µL of MMLV RT), RNA template (0.4 - 0.5 µg/µL), and sterile deionized water. The reaction was performed at 25oC for ten minutes, at 48oC for 30 minutes, and at 94°C for five minutes.
3.2.3. Polymerase Chain Reaction Assay
For the polymerase chain reaction (PCR) assay, primers were designed for CDV and B. bronchiseptica with the following sequences: CadF: AAGCCTCACACTGTTCAAG-3’, CadR: GATTAGGACTATAATGACATGC-3’; and BbroF: TGGCGCCTGCCCTATC-3’, BbroR: 5’- AGGCTCCCAAGAGAGAAAGGCTT-3’. The PCR assay was carried out at 95°C for four minutes in one cycle, at 95°C for 30 seconds in 30 cycles, at 67°C (for B. bronchiseptica) or 57°C (for CDV) for 30 seconds, and at 72°C for 30 seconds. A final extension was also performed at 72°C for four minutes. The positive and negative control samples, prepared by Isfahan University of Medical Sciences, were also included in each test. Next, gel electrophoresis of PCR products was carried out for both controls and test samples in 1.5% agarose stained with DNA green viewer. Finally, the positive samples were sent to Bioneer Co., (Korea) for sequencing.
4. Results
4.1. Rapid Immunochromatography
A total of 29 positive samples were found among 50 eye samples and respiratory swabs taken from dogs with suspected symptoms of DCV infection, using a rapid immunochromatography kit. Also, one positive sample was found among 50 eye samples and respiratory swabs taken from seemingly healthy dogs.
4.2. PCR and RT-PCR Assays
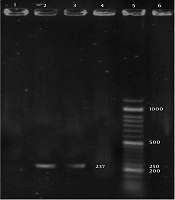
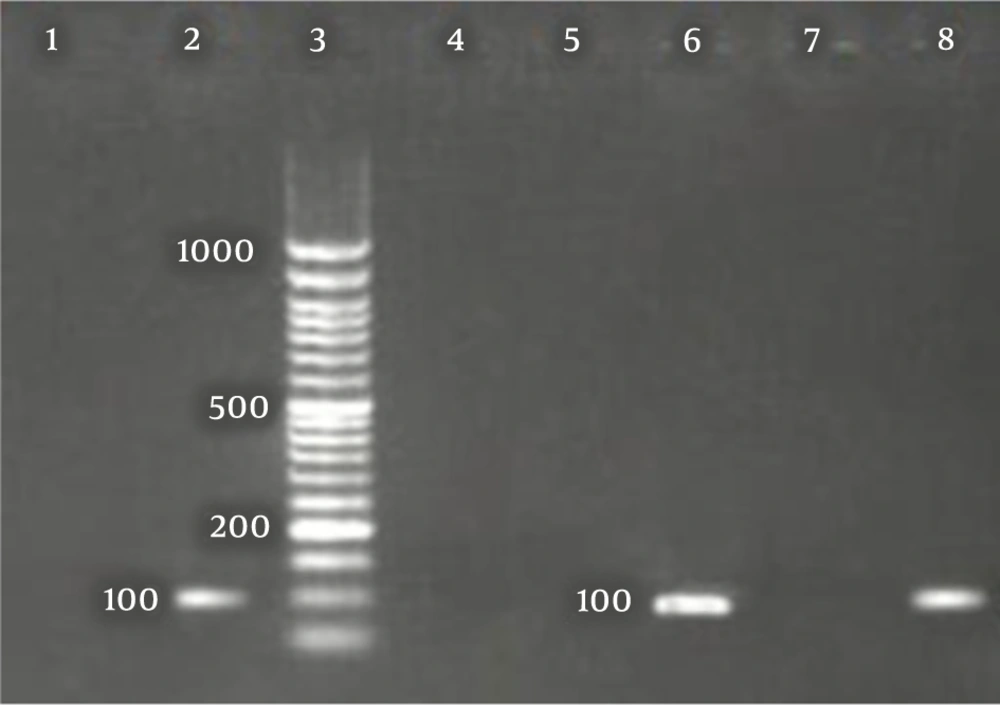
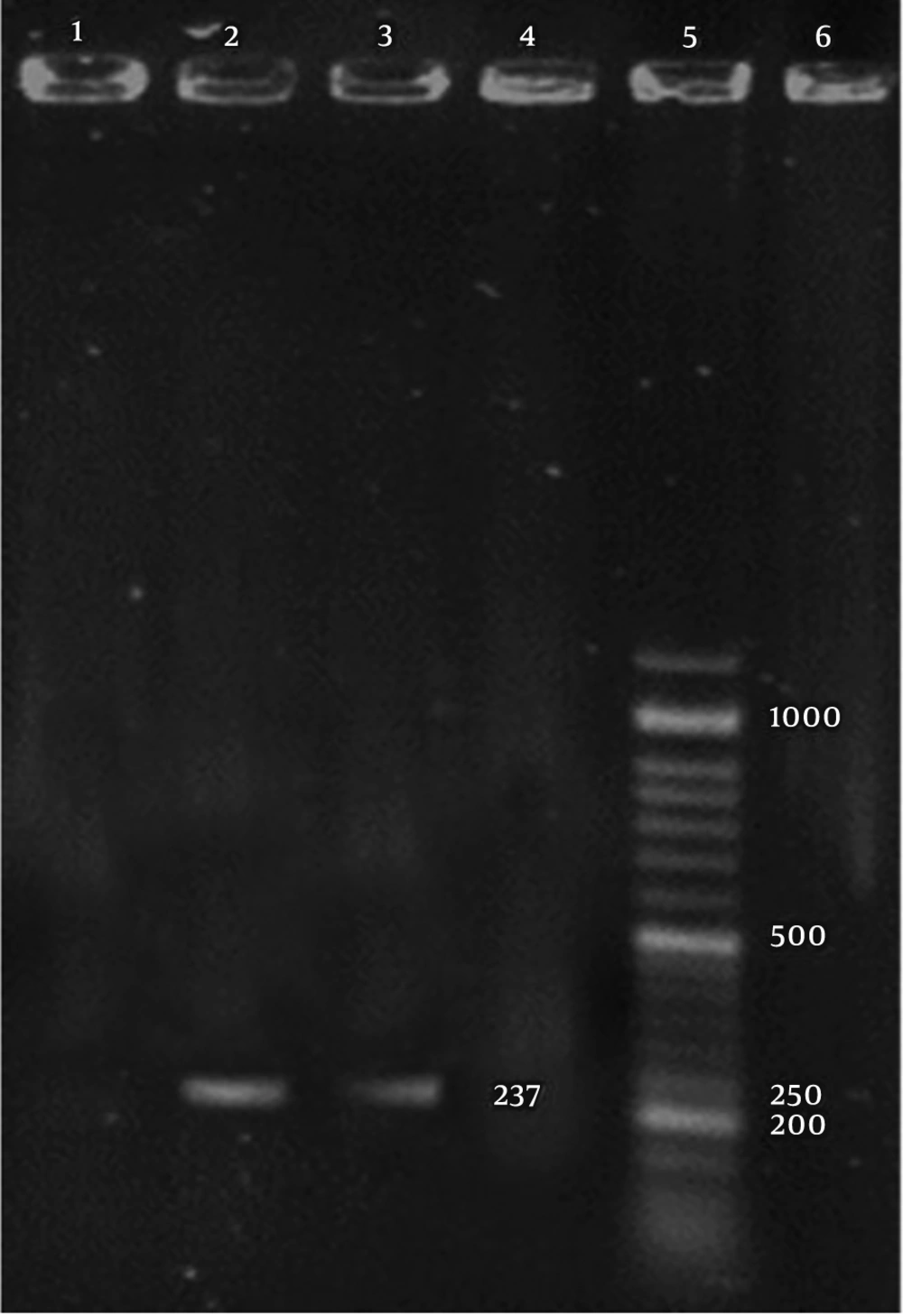
The CDV and B. bronchiseptica- specific bands with sizes of 100 and 237 bp, were detected in the positive controls and positive PCR products, respectively (Figures 1 and 2).
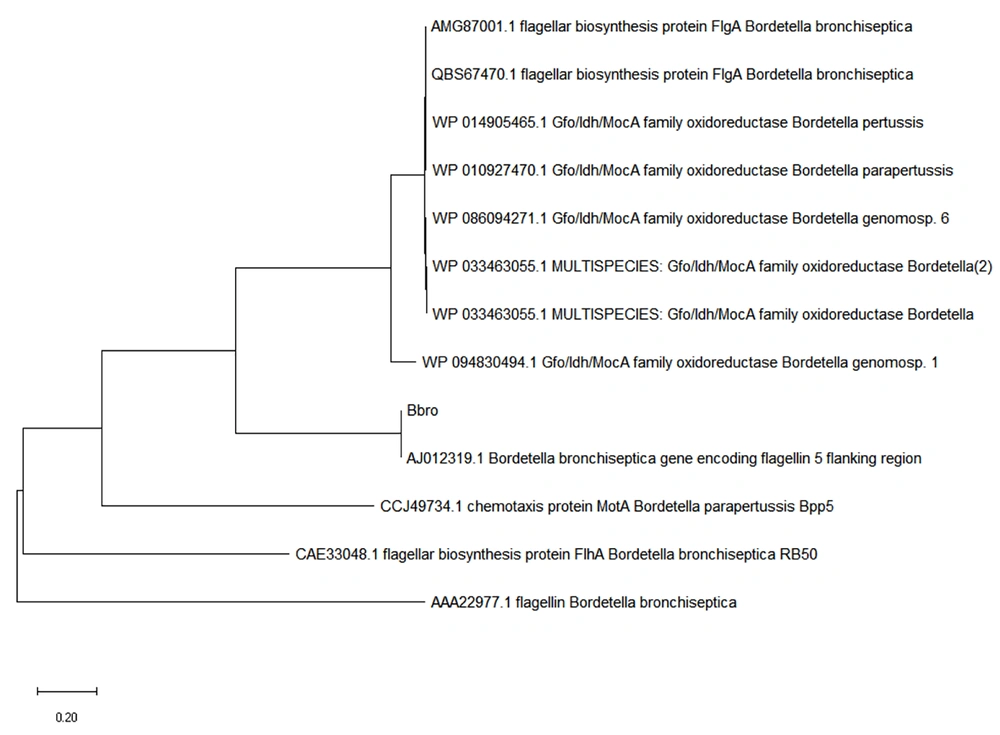
The degree of homology between the sequence extracted from the PCR product reading of the studied genes and the sequences registered in the GenBank for CDV and B. bronchiseptica was a proper indicator of PCR adequacy (Figures 3 and 4).
The evolutionary history was inferred using the neighbor-joining method; the optimal tree is shown here. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and represented the number of amino acid substitutions per site; this analysis involved 26 amino acid sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion). There were 524 positions in the final dataset. All evolutionary analyses were conducted using the MEGA X tool.
The evolutionary history was inferred using the Neighbor-Joining method; The optimal tree is shown here. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and represented the number of amino acid substitutions per site. This analysis involved 26 amino acid sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion). There were 524 positions in the final dataset. All evolutionary analyses were conducted using the MEGA X tool.
Out of 50 samples taken from dogs with suspicious CDV, 37 positive samples were detected for the presence of CDV nucleic acids using RT-PCR. Also, out of 50 samples taken from seemingly healthy dogs, three were found to be positive for CDV nucleic acids. Based on the results of PCR assay for B. bronchieptica, out of 50 samples taken from dogs with suspected CDV clinical signs, 20 were positive for the presence of B. bronchieptica nucleic acids. Also, out of 50 samples taken from seemingly healthy dogs, one sample was positive for the presence of B. bronchieptica nucleic acids.
4.3. Statistical Analysis
After entering the data in SPSS version 16 and drawing a two-way agreement table, the relationship between variables was examined, using the Chi-square test. However, no significant relationship was found between the variables (P > 0.05).
5. Discussion
Almost all dogs with canine distemper have lymphopenia and a weakened immune system; therefore, other infections can accompany this disease and compromise the animals’ health (17, 18). Immune-suppressing mechanisms in cells may be involved in canine distemper that are not directly associated with viral infections. All researchers and scholars confirm the weakening immune system of animals due to CDV. The present study was performed to identify infections with CDV and B. bronchiseptica as another respiratory pathogen in dogs. Despite the simultaneous detection of these two pathogens in 4% of samples, there was no significant association between CDV and B. bronchiseptica.
The specificity and sensitivity of immunochromatography for samples obtained from the conjunctiva are similar to nested PCR (100%), because when the viral disease is in the acute phase, the virus rapidly infects the lacrimal glands and conjunctival secretions; vaccination also does not interfere with the results of this test (18, 19). Therefore, in the present study, this method was used for the initial diagnosis of distemper.
So far, several researchers have investigated the etiology of canine respiratory infections and their concurrence. In 2015, Lavan et al. (5) conducted a study on the frequency of some respiratory agents in seemingly healthy dogs from animal shelters in the United States. In this study, M. cynos (29.2%), B. bronchiseptica (19.5%), canine adenovirus type 2 (CAV-2) (12.5%), CDV (7.4%), and canine parainfluenza virus (CPIV) (3.2%) had the highest frequencies of all identified pathogens. This study found that the prevalence of B. bronchiseptica infection was higher than CDV in asymptomatic dogs, which is consistent with the present findings. Also, in the current study, the prevalence of B. bronchiseptica in apparently healthy dogs was higher than that reported in the study by Lavan et al. (5) (30% vs. 19%). Factors, such as the dog’s living environment (e.g., temperature, hygiene, solitary or herd, and vaccination), may contribute to differences in the prevalence of this pathogen.
Furthermore, Headley et al. (cited in Ford) described canine distemper along with secondary pneumonia due to B. bronchiseptica in two mongrel dogs (20); however, this study did not investigate the prevalence of infection and only reported coinfections. The possibility of coinfection with CDV and B. bronchiseptica has been reported in the literature. In 2014, Schulz et al. (21) detected a number of viruses, as well as B. bronchiseptica, in symptomatic dogs. In their study, nasopharyngeal samples were examined for CAV-2, canine influenza virus (CIV), CRCoV, CHV-1, CDV, and B. bronchiseptica nucleic acids by PCR assays. Overall, 37.7%, 8.8%, and 78.7% of dogs with canine infectious respiratory disease (CIRD) were positive for CPIV, CRCoV, and B. bronchiseptica, respectively. Concurrent diseases were identified in 47.9%, 82.6%, and 100% of animals positive for B. bronchiseptica, CPIV, and CRCoV, respectively. Among asymptomatic animals, 1.1%, 7.8%, and 6.45% were positive for CAV-2, CPIV, and B. bronchiseptica, respectively. Infection with CPIV and B. bronchiseptica in dogs with CIRD was notably more frequent than asymptomatic dogs (21). In the present study, the prevalence of B. bronchiseptica in asymptomatic cases was 40% lower than the rate reported by Schulz et al. (21) in 2014. Possible differences in the dogs’ living environment and their breed are among factors influencing this difference (22).
Moreover, Decaro et al. (9) in 2016 performed a molecular study to detect the etiology of CIRD among animals with clinical CIRD, healthy dogs, and animals with the preliminary stages of CIRD. CPIV had the highest frequency among CIRD cases, followed by CRCoV, B. bronchiseptica, M. cynos, M. canis, and pneumovirus. In the present study, the frequency of B. bronchiseptica was significant, whereas the frequency of CDV was much higher than the study by Decaro et al. in 2016 (9). Since in the present study, we aimed to determine the relationship between CDV and B. bronchiseptica and conducted non-random sampling, the difference in the frequency of these viruses between these two studies can be justified.