1. Context
Primary bone tumors are relatively uncommon, generally accounting for 0.2% of all malignancies and 0.3% of cancer deaths (1). Bimodal age distribution with two peaks of incidence is observed in bone tumors, the first of which is noticed in childhood/adolescence, especially 10–14-year-old aged patients, and the second of which is reported in late adulthood, especially individuals aged above 65 years and frequently suffering from Paget's disease (2). Bone tumors were classified according to the differentiation of malignant cells based on a list of parameters such as cell morphology, phenotype, and genotype (1).
Except for chondrosarcoma, osteosarcoma (OS) is the most frequent primary bone tumor, accounting for about 16 - 17% of bone tumors (1). According to the literature, OS comprises several tumors with slightly different clinical, radiological, and histological features (2-4). Moreover, OS affects 5/106 persons per year and represents the most diffused primary bone tumor among children (5), accounting for 5% of cancers and 8.9% of cancer-related deaths among these individuals (6). Differences in its prevalence have also been reported regarding gender, with annual prevalence rates of 5.4/106 and 4/106 among males and females, respectively (6).
Several abnormalities are described in OS, making it difficult to define its diagnosis, select an appropriate therapy, and formulate the prognosis (7). The recognition of specific OS variants is critical for the treatment of the patients (8). Epidemiology data, genomic/molecular characterization, and studies in experimental models have provided helpful insights to OS management. Although many research efforts have been made, the aetiology, pathogenesis, and natural history of OS are not well detected.
The present review study aimed to resume and discuss the state of the art in the role of selected signal transduction pathways in the emergence of Osteosarcoma and during metastasis formation.
2. Evidence Acquisition
2.1. OS Features
OS seems to originate from osteoid-producing mesenchymal stem cells (MSCs), located in multiple compartments (7, 9). MSCs include adherent cells in the intramedullary space and osteoblasts located along bone endosteum and periosteum, which are actively involved in fracture repairs and remodeling (10, 11).
Malignant OS cells produce osteoid or immature bone (2, 3), resulting in lesions that grow radially form spheroidal masses. Such neo-formations penetratethe bony cortex into a capsule-like layer, referred to as the reactive zone, compressing the neighboring muscles. The extensions of the primary tumor, i.e., satellites, invade the reactive zone. The tumor invasion induces bone weakness, pain, swelling, tenderness, and fractures in the affected area. Occasionally, lethargy, fever, weight loss, and anemia are also observed (12).
Although OS can virtually develop in any bone, the primary tumor mostly triggers in the actively-growing metaphyses of long bones (e.g., distally in the femur and proximally in the tibia and humerus). Differences in the tumor site and patients’ survival rates seemingly depend on the age at diagnosis (13, 14).
The tumor can diffuse regionally or spread to other organs (15). About 20% of patients are referred to clinics/hospitals with lung metastases at the moment of diagnosis. Otherwise, metastases can occur in about 40% of patients with nonmetastatic OS (12). The OS metastases primarily colonize the lungs, and usually, after metastasis has formed in the lungs, then metastases can occur in bones different from the primary tumor. Metastases in distant bones represent the last OS stage and are associated with the poorest prognosis (12, 16). From this perspective, the metastatic spreading is considered as a crucial aspect of the OS progression.
The OS recognition is complex, and it is not often possible to make the diagnosis exclusively by radiologic examination. Histology observations and the consistent spectrum of clinical presentations help formulate the diagnosis, contributing to the so-called triple diagnostic approach, which is necessary to discriminate different OS features.
Depending on the features of tumors, the OS treatment includes a combination of surgeries, radiotherapy, and chemotherapy. In this regard, the metastases aggravate the prognosis (2), as metastasizing lesions partially respond to therapies and are the leading cause of death (4). Accordingly, both the progression of the disease and the outcomes mainly depend on the metastatic potential of OS, which has a high tendency to disseminate (2, 3).
2.2. Formation of Metastasis and Signaling
To develop a metastasis, the malignant cell must leave the primary tumor site, migrate, survive within vessels, invade, and colonize a secondary site. The molecular evolution of cancer cells, while they are spreading, is still in need of research as such metastases still are considered as a challenge for personalized cancer therapy. Recently, the concept ‘metastasome’ has been suggested to revise the metastasis model and systematize both molecular and functional features characterizing metastases differently from primary tumors (17, 18).
The development of metastases is a complex phenomenon with some sequential steps, and the acquisition of motility is a central event for the malignant cell (5, 19-21). The sequence of events ultimately enabling cells to migrate results in the reorganization of the cytoskeleton, which involves several signal transduction pathways such as the Rho/Rac GTPase signaling (22, 23). The Rho/Rac GTPase pathway induces membrane ruffles and is involved in forming focal adhesion complexes and lamellipodia and prompting cells to extend the protrusions requested for movement (24). In invasive tumor cells, the components of signaling pathways in the cytoskeleton architecture are abnormally expressed (20, 21). Despite research efforts, the intracellular mechanism regulating cell migration is not well-understood.
The knowledge of the signal transduction systems acting in the bone tissue, which can be dis-regulated during the OS emergence and progression, might provide promising perspectives. The detection of molecules belonging to signal transduction pathways might contribute to describing the natural history of the tumor and refine the prognosis, opening new ways to novel molecular-targeted therapeutic strategies.
From this perspective, Ezrin, a protein involved in regulating the cytoskeleton activity and the metastatic spread of OS, has been of great interest (25).
2.3. Ezrin and OS Metastases
Ezrin belongs to the family of ERM proteins, which play structural and regulatory roles in the assembly and stabilization of specialized plasma membrane domains by cross-linking plasma membrane proteins and actin filaments (26). As a member of the ERM family, Ezrin possesses the Protein 4.1 ezrin, radixin, moesin (FERM) domain (PBD ID:1GC6) (27). The N-terminus FERM domain is demonstrated to interact with membrane/transmembrane proteins and signaling molecules (27-29). Ezrin participates in complex cell surface structures and is involved in some activities, including the Rho GTPase-dependent signal transduction pathway.
The imbalanced inactive versus active conformation of the ERM proteins plays a key role in the conformational regulation of the link between cytoskeleton actin and the plasma membrane (29, 30). The Ezrin activation is strictly regulated, mainly depending upon the phosphorylation/de-phosphorylation imbalance. Ezrin undergoes phosphorylation at two sites: (1) C-terminal threonine (Thr 567) using enzymes bearing kinase activity such as Rho kinase (31), and (2) N-terminal tyrosines by means of receptor tyrosine kinases such as the epidermal growth factor receptor and c-MET (32, 33). This phenomenon promotes the reorganization of the cytoskeleton, inducing morphogenetic and functional modifications in the cell. The un-phosphorylated Ezrin, located in the cytoplasm, can be activated by phosphorylation and moves and tether actin to the cortical membrane (34).
Evidence indicates that the absence of Ezrin (both expression and function disruption) decreases the OS spreading. In experimental models, a significant decrease in the metastatic dissemination was obtained after suppressing Ezrin’s expression or disrupting its function by the use of antisense transfection, the stable expression of short hairpin RNA, or the transfection of a dominant-negative Ezrin (25, 26). The absence or reduction of Ezrin was associated with a decrease in Akt and mitogen-activated protein kinase (MAPK) activity (35). Moreover, the imbalanced activation/inactivation of the ERM proteins indirectly depends on the calcium metabolism (36).
2.4. Ezrin and Calcium Signaling
The calcium concentration regulation is critical to many physiological events; hence, a finely-tuned regulation by a complex network of signaling pathways is required. Intracellular calcium plays a crucial role as a second messenger in regulating cell apoptosis (37). Abnormal calcium increase within the cell affects protein functions, secretion, and contractility (37).
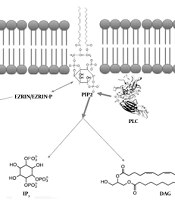
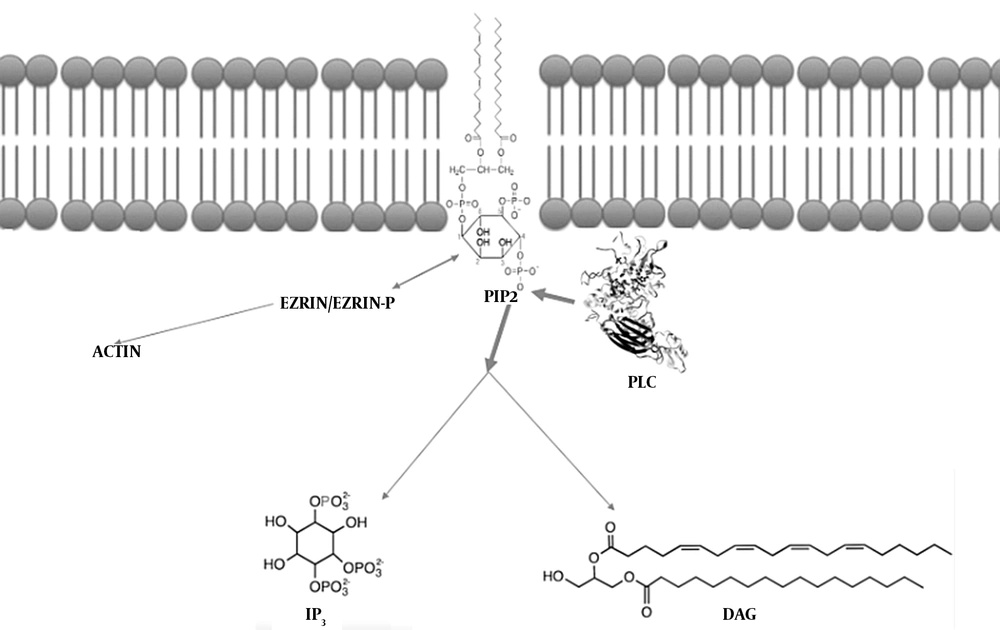
Calcium metabolism is regulated by the signaling pathways, including the phosphoinositide (PI) transduction system (38). The phosphorylated PI derivative phosphatydil inositol (4,5) bisphosphate (PIP2), located in the inner half of the membrane’s lipid bilayer, is of essence for many cellular activities, including endo/exocytosis, cell motility, and ion channel regulation. The PIP2 levels are strictly regulated by converting enzymes (39), including the PI-specific phospholipase C (PLC) family (37, 39). Great attention has recently been paid to the PLC enzymes due to their involvement in an increasing number of diseases (40-43)
The calcium metabolism was demonstrated to be critical for the viability of human OS cell lines (2, 3) and the relationship between Ezrin and PIP2. Depending on the N-terminal domains, the ERM proteins can simultaneously bind both actin and PIP2 (44-49). The conformation of Ezrin is changed after it interacts with PIP2. The change in Ezrin’s conformation activates ezrin (47). In this regard, both phosphorylation and PIP2 binding allow the stabilization of ERM proteins and/or ameliorate binding to receptors (49-52).
As the Ezrin activity depends on membrane PIP2 levels, PIP2-cleaving PLC enzymes play a critical role. PLCs are required for the chemokine-mediated dissociation of ERM proteins from the membrane (53). The decreased PIP2 levels move ERM proteins to dissociate from the plasma membrane. The inactivation follows the chemokine binding to a GPCR, CXCR4, thus supporting the hypothesis indicating that the PLC activation commonly results from GPCR signaling (38).
2.5. The PI-specific PLC
The activation of PLC basically cleaves the polar head group of PIP2, producing two further signaling molecules (namely inositol trisphosphate (IP3) and diacylglycerol (DAG)) (39).
As IP3 is water-soluble, it rapidly diffuses to the cytoplasm, binds IP3-gated calcium channels in the endoplasmic reticulum (ER) membrane, and induces calcium release from the ER. The calcium increase in the cytoplasm initially propagates as a wave, followed by a series of calcium spikes (39).
Binding to the membrane by using its fatty acid tails, DAG can be cleaved into arachidonic acid to synthesize eicosanoids (54) and activate serine/threonine calcium-dependent protein kinase C (PKC) enzymes. Moreover, the increased IP3-induced calcium moves PKC from the cytoplasm to the cytoplasmic face of the plasma membrane, thereby activating and enabling PKC to phosphorylate further target proteins (37, 39, 55).
The mammals' PLC family comprises multi-domain enzymes grouped into six sub-families based on the amino acid sequences, domain structures, and recruitment mechanisms: β(1-4), γ(1-2), δ(1, 3, 4), ε(1), ζ(1), and η(1-2) (37).
The PLC enzymes, specifically PLC ζ, which is exclusively described in the spermatic lineage (37), are cell-type specific (56-58). The panel of PLC expression in each cell varies under abnormal conditions, compared to the normal counterparts (43, 59, 60). The panel of PLC expression was also modified by specific stimuli (61-63), including inflammation (64-66), or by some specific inhibitors (67, 68). The subcellular distribution of the PLC enzymes influences their activity, suggesting specific roles for each isoform besides the cleavage of PIP2 (69-73). The PLC enzymes are involved in some human diseases, including the nervous system tumors (64, 65) and abnormalities (64, 74-79).
2.6. PLCs in the Bone
The PLC enzymes contribute to a broad spectrum of regulatory interactions with receptor and non-receptor tyrosine kinases and the lipid components of cellular membranes in all human tissues (37). A review of the literature indicates how PLCs can specifically act in the complex bone metabolisms. In this regard, the activation of PLC is important in modulating the parathyroid hormone (PTH) (80). PLC β1 isoform might commit MSCs to differentiate into osteogenic lineage cells (81, 82). The interaction between the calcitonin gene-related peptide (CGRP) and its own receptor activates PLC β1 (83). Berberine hydrochloride targets TRAF6 and NFATc1, inhibiting osteoclastogenesis, and bone destruction also decreases the PLC γ1 levels (84). PLC γ2 is demonstrated to be a key element in the early phases of osteoclast differentiation, adhesion, and migration, thus playing a role in bone resorption (85, 86). During autoimmune inflammation, PLC γ2 probably contributes to regulating the cellular and molecular mechanisms in bone and immune cells (87). Furthermore, PLC γ2 regulates the activation of Proto-oncogene tyrosine-protein kinase Src (c-Src) and membrane localization in osteoclasts (88).
Although reports have suggested that the PLC enzymes may represent critical regulators in the bone adaptation, their role is not well-explored.
2.7. PLCs in Human Osteoblast
Osteoblasts derive from MSCs and play a pivotal role in secreting the matrix components of bones (89-91). Depending on many factors, the osteoblast can differentiate into osteocytes (92) or chondroid bone-forming cells under specific conditions (93).
In osteoblasts, the selected PLC enzymes contribute to regulating the calcium metabolism in a complex manner (94-99). Following the mechanic stimulation, the nuclear factor kB (NFkB) moves into the nucleus, under the PLC control (100, 101).
In osteoblasts, a peculiar panel of the PLC expression is reported (102). In this regard, PLCB1, PLCB3, PLCB4, PLCG1, PLCG2, PLCD1, PLCD3, PLCD4, and PLCE are expressed, while PLCH1 and PLCH2 are inconstantly expressed (Table 1). Moreover, PLC β1, PLC β4, PLC γ1, PLC δ1, and PLC η1 enzymes are detected in the cytoplasm. PLC β3 and PLC η2 are detected at the perinuclear and membrane levels, respectively (102). Despite its absence, PLC β2 in osteoblasts transduces signals from prostaglandin E2 and other prostanoids (103). Given the extremely low basal expression, the PLC β2 expression seems to be correlated with specific stimuli, including transforming signals (103), as discussed above.
| OMIM | Fibroblast | Osteoblast | MG-63 | SaOS-2 | Hs888 | 143B | |
|---|---|---|---|---|---|---|---|
| PLCB1 | *607120 | + | + | + | + | + | + |
| PLCB2 | *604114 | - | - | - | - | - | - |
| PLCB3 | *600230 | + | + | + | + | + | + |
| PLCB4 | *600810 | + | + | + | + | + | + |
| PLCG1 | *172420 | + | + | + | + | + | + |
| PLCG2 | *600220 | + | + | + | + | + | + |
| PLCD1 | *602142 | + | + | + | + | + | + |
| PLCD3 | *608795 | + | + | + | + | + | + |
| PLCD4 | *605939 | + | + | - | - | + | - |
| PLCE | *608414 | + | + | + | + | + | + |
| PLCH1 | *612835 | - | + | - | + | + | - |
| PLCH2 | *612836 | - | + | - | - | - | - |
Although the PLC δ4 enzyme is not detected and a PLCD4 transcript is expressed in osteoblasts and the Hs888 metastatic cell line, it is absent in other OS cell lines, including MG-63 and SaOS-2 cells (102). Interestingly, PLCD4 was inconstantly expressed in fibroblasts (58). The sub-family isoforms of PLC δ, the most evolutionary ancient, and well conserved (37), are highly sensitive to calcium and play a key role in cell proliferation (37). The PLC δ4 involvement is observed in response to nutritional and environmental stress in yeast and plants (104, 105), and in stress-induced response in human endothelial cells (61, 62, 65, 102). Compared to normal resting liver, PLC δ4 is expressed at high levels to regenerate liver in hepatoma and src-transformed cells (106). Accordingly, PLC δ4, in cooperation with nuclear PKC α and ε regulates liver regeneration (107). The PLC δ4 expression is probably associated with mitogenic signals, so this isoform is expressed more abundantly in high-rate proliferating cells (108), as discussed above. The up-regulation of PLCD4 might regulate other PLC enzymes, including PLC β1 (109). This issue might be due to the highest sensitivity of PLC δ isoforms to calcium concentration. Probably, the PLC δ4 activation by a small amount of calcium induces an increase in the calcium levels to recruit further PLC isoforms, implying an internal regulatory hierarchy among the PLC enzymes.
Similarly, the PLC η isoforms, the most recently discovered PLC family, show high sensitivity to calcium (37, 110). PLC η1 as a signal amplifier plays a key role in calcium signaling mediated by G protein-coupled receptors (GPCR). The mobilization of ER intracellular calcium plays a pivotal role in the PLC η1 activation (111), which is probably caused by a positive feedback from PLC δ4 (66, 68). On the other hand, PLC η2 is involved in regulating temporal and spatial cell calcium dynamics (110), thus transducing calcium signals from mitochondria. Osteoblasts, MG-63 cells, and SaOS-2 cells seem to share a similar expression panel of the PLC enzymes (65), with significant differences in expressing the PLC δ4 and PLC η subfamily isoforms.
2.8. PLC in Human OS Cell Lines
Previous reports analyzed the expression of PLCs in human bone tumors (68), with particular attention to OS. This is because of the relationship between Ezrin and PLC, which can interact with PIP2. The OS cell lines with different molecular, histologic and clinical features have been extensively researched (63, 65, 68, 77, 109).
As partially expected, the expression panel of PLC enzymes depends on a specific OS cell line (65). The MG-63 cells have low levels of alkaline phosphatase activity and PTH unresponsive adenylate cyclase (112). Although the MG-63 cell line is usually used as an experimental model for human osteoblasts, the expression panel of PLCs differs from the panel described in osteoblasts, as in the other OS cell lines (63, 68). The specific features of each OS cell line partially explain different responses to therapy, prognosis, and expression panel of PLCs. SaOS-2 epithelial-like cells are also used as an experimental model for human osteoblasts (100, 113, 114). In this regard, 143B cells, thymidine kinase negative, are considered highly aggressive and develop osteolytic tumors (115). Hs888 cells are derived from OS lung metastases (25).
Previous studies have indicated the expressions of PLC β1, β3, γ1, γ2, and δ1 in some osteosarcoma cell lines, including MG-63 and SaOS-2 cell lines (116, 117). More recent studies have suggested that the MG-63 cell line expresses PLC β1, β2, β3, γ1, γ2, β4, δ3, and δ1 (65). In the143B cell line, PLC β1, β3, β4, γ1, γ2, δ1, δ3, and ε are also expressed; however, PLC β2 is weakly expressed. In the SaOS-2 cell line, PLC β1, β3, γ1, γ2, δ1, β4, δ3, ε and η1 are expressed, and PLC β1, β3, β4, γ1, δ1, δ3, δ4, and ε are expressed in the Hs888 cell line (65) (Table 1).
PLC β2, involved in mechanic transduction and cell attachment, inconstantly described in osteoblasts (61, 103), was weakly expressed in the MG-63 cell line (65). PLC β2 was not described in unstimulated cells (61, 116, 117), suggesting that the basal expression is low, and increase might follow to different and specific stimuli, both mechanic and biochemical (103).
Enzymes belonging to the PLC γ subfamily are highly expressed in tumors compared to normal tissues (118, 119), thereby playing key roles in cell growth, survival, and migration (120). Rho/Rac GTPases contributes to the cytoskeleton reorganization, thus enabling cells to extend the protrusions requested for movement (22-24). PLC γ1, a critical factor to develop and maintain tumor metastases, is noticed in four human OS cell lines (65). PI3 kinases (PI3Ks), together with Rac, regulates both polarization and cell migration (24), contributes to activating PLC γ1 (121) and translates to the leading edge of MDA-MB-231 migrating cells (117). In this regard, PLC γ1 probably acts as an intracellular link between PI3K and Rac and regulates the reorganization of actin by the Rac and WASP family proteins (122). However, PLC γ1 can regulate cell migration by further downstream effectors and directly activates cofilin (123), which is also involved in the motility and invasion of malignant cells. PLC γ1 can regulate cell migration and invasion by modulating the activation of ERK acting as a downstream effector (21). Moreover, the small interfering RNA knockdown of PLCG1 in human prostate carcinoma cell line (PC3LN3) affects cell adhesion (124). In breast cancer, the dissemination of malignant cells is correlated with the PLC γ1 expression, and the absence of the enzyme affects actin polymerization and focal adhesion modulating Rac (122). In addition to the critical role of PLC γ1 in the metastasis formation, its down-regulation results in metastases regression (122). Phospholipase C gamma1 is required for metastasis development and progression, explaining the regression of micrometastasis.
PLC γ2 is expressed in MG-63, 143B, and SaOS-2 cell lines; however, it is exclusively absent in Hs888 cell line (65). In this regard, its concentration is estimated to be about 50% higher in 143B and SaOS-2 cells than in MG-63 cells (65). This may be justified based on the aforementioned role of PLC γ2 in bone resorption during the differentiation of osteoclasts (85-87).
PLC δ4 has been expressed exclusively in Hs888 (65) and in different tumors (40, 56, 64). In breast cancer cells, the abnormal expression of PLC δ4 induces both the up-regulation of ErbB and the activation of the ERK pathway (125).
Regarding their features, Hs888 cells express PLC δ4 but not PLC γ2 (65). PLC η1, expressed in SaOS-2 and Hs888 cell lines (65), is essential for downstream ERK1/2 phosphorylation (111). Although the role of PLC η1 is not well-investigated, its activation seems to enhance the GPCR-mediated calcium pathway (111).
2.9. Ezrin and PLCs in OS
The abnormal expression of Ezrin is associated with the poor prognosis in human tumors, suggesting its role in tumor progression (126). Ezrin induces metastatic behavior in animal OS models (25), which may interfere with apoptosis, cell adhesion, and/or Rho GTPases signaling (127). PLCs induce ERM proteins to dissociate from the membrane by reducing the PIP2 levels (53). Inactivation follows chemokine’s binding to CXCR4, suggesting that PLC activation is the outcome of the GPCR signaling (38). Enzymes belonging to the PLC β sub-family are activated by Rac, and PLC ε is a downstream effector of Ras superfamily GTPases and an upstream activator of small GTPases, including both Ras and Rap (128) as key regulators in actin cytoskeleton rearrangement and tumor cell migration and invasion (129-131). PLCs probably act as the convergence point of the signaling pathways networking Rho and Ras GTPases, which in turn regulate the Ezrin activity (36, 132). From this perspective, a relationship between Ezrin with the PLC enzymes and the metastatic progression of OS was hypothesized.
Given the supposed role of Ezrin in tumor progression and metastasis formation, silencing the expression of EZR (OMIM *123900), a gene codifying for Ezrin protein, significantly reduces the growth rate of both 143B and Hs888 cells in a time-dependent manner, with the most marked effects observed in the 143B cell line (61).
In the 143B cell line, the EZR silencing and the consequent lack of Ezrin induce the up-regulation of PLCG2, the down-regulation of PLCB1 and PLCE, and cell morphology changes and reduce intercellular adhesion and cytoplasm microvacuolization (109).
In the Hs888 cell line, EZR silencing induces the up-regulation of PLCB1, PLCG2, and PLCD4, and cell morphology changes such as decreasing the number of cells with well-preserved structure. In the un-silenced control Hs888 cells, PLC β1 is mildly detected in the cytoplasm, while PLC γ2 is absent. Following the Ezrin silencing, β1 significantly increases in the cytoplasm PLC, and PLC γ2 is weakly detected, mainly evident in the perinuclear region (109).
In the 143B cell line, EZR silencing reduces, while it enhances the transcription of PLCB1 in Hs888. Notably, in both 143B and Hs888 cell lines, PLCB1 is expressed at a low rate (65). The expression of PLC β1 is selectively increased during myoblast and adipocyte differentiation (133, 134) and development (135) in breast cancer (120, 136). In high risk patients affected with myelodisplastic syndrome, the deletion of PLCB1 is associated with poor outcome progressing to acute myeloid leukemia (137, 138). In this regard, as PLCB1 loss favours myeloid lineage cancer (42, 137), it is speculated that normal/increased PLCB1 may be a disadvantage to malignant progression. The loss of Ezrin, which promotes metastasis formation, induces the down-regulation of PLC β1 in the most aggressive OS cell line, which may favour malignancy. The variations in the PLCB1 expression, which concomitantly occur with the Ezrin silencing, suggest a relationship between the two molecules. This issue deserves further studies.
The EZR silencing induces the up-regulation of PLCG2 transcription (109). The PLC γ sub-family isoforms bear a unique region comprising two tandem SH2 domains and one SH3 domain adjacent to a split Pleckstrin Homology (PH) domain (139, 140). Notably, Ezrin interacts with the SH2 domain, and reveals a relationship with PLC γ sub-family (34). PLC γ2 is involved in the actin cytoskeleton reorganization and Rac-activation in dendritic cells (141). As previously discussed, PLC γ2 plays a critical role in the osteoclast lineage, acting in the integrin-mediated adhesion and migration (85), and in the activation of Proto-oncogene tyrosine-protein kinase Src (c-Src) and membrane localization (88). Actin binding proteins, which bind PIP2, negatively regulate PLC γ2; hence, the up-regulation of PLC γ2 following the EZR silencing might be associated with the osteolytic nature of 143B cells as PLC γ2 acts in osteoclast formation and function (88) as well as in bone loss modulation (141). In this regard, PLC γ2 is not detected in Hs888. After EZR silencing, the transcript from PLCG2 is detected, indicating that the regulation of PLC γ2 networks Ezrin, independently from the OS cell line.
PLC δ4, exclusively expressed in the Hs888 OS cell line, is up-regulated after the EZR silencing. In breast cancer cells, the abnormal expression of PLC δ4 contributes to carcinogenesis by up-regulating ErbB and activating the ERK pathway (125) in response to mitogenic signals (108). The up-regulation of PLCD4 in Hs888 following the EZR silencing suggests a relationship between Ezrin and PLC δ4, which seems to be associated with the contemporary up-regulation of PLCB1 (109, 142).
In Hs888, the EZR silencing induced significant differences in the expression of PLCE (109). PLC ε plays a key role in carcinogenesis, with an undefined mechanism of action. The experimental loss of PLC ε inhibits the growth of bladder tumor cells (143, 144), and the over-expression of PLC ε is reported in murine skin cancer (145-147). PLC ε is involved in the progression of head and neck cancer (148), and one PLCE polymorphism is associated with squamous cell carcinoma and gastric cancer (36). Controversial reports have indicated both tumor-suppressing and promoting activities for PLC ε, which may depend on the type of tumor. Considering OS, the Ezrin silencing enhances the expression of PLCE in the Hs888 cell line, suggesting a relationship between Ezrin and PLC ε. Microscopy observation confirmed this hypothesis, suggesting that Ezrin and PLC ε focally and partially co-localize in the cytoplasm of the OS cells (109).
Remarkably, in the OS cell lines, the EZR silencing modifies the expression of PLC genes in a complex manner (109). The overall reorganization of the expression panel of PLC genes following the EZR silencing indicates a functional connection between PLC and Ezrin. Further studies on the relationship between Ezrin and selected PLC enzymes may elucidate the crosstalk of signal transduction pathways in OS.
2.10. Ezrin, PLC, and Ras GTPases
The selected PLC enzymes represent the convergence point of pathways promoting Rho and Ras GTPase-mediated signaling, which in turn regulates the Ezrin activation. Among the wide spectrum of regulatory interactions covered by the PLC family, theere has been a great interest in direct binding to Ras GTPases (37-39, 149-151) regarding isoforms belonging to the PLC β and ε sub-families.
The PH domain in the structure of enzymes belonging to the PLC β sub-family interacts with the Rho-GTPase family member Rac, thus activating PLC (36). Rac1, involved in actin polymerization, cell cycle regulation, and proliferation (152-155), can induce in vitro malignant transformation of the cell (156, 157). PLC ε is a direct effector for Rho GTPases (158), directly and concomitantly activated by both RhoA and individual Ras GTPases. Activation caused by a variety of hormones, neurotransmitters, and growth factors results in the upstream control of signal transduction pathways in the PLC ε downstream (37, 109).
Accordingly, isoforms belonging to the PLC β and ε sub-families as well as Ezrin undergo Ras signaling regulation. On the other hand, both Ezrin and PLC recognize and may competitively bind plasma membrane PIP2 molecules. A decrease in PIP2 induces Ezrin to relocate in the cell (159); however, the inactivation of RhoA inhibits such relocation, suggesting that RhoA might be involved in the pathway upstream by either masking the actin binding site (160) or using a common downstream effector (160). The expression of dominant negative Rac1 reduces Ezrin at the level of adherens junctions (159).
The expression and the sub-cellular distribution of RhoA and Rac1 change after silencing both EZR and PLCE genes (142). Further, the PLC ε silencing acts differently on the 143B and Hs888 cell lines depending on the different features of the cells (142). The perinuclear relocation of Ezrin was associated with a decrease in RhoA and Rac1, which may be caused by the PLC ε loss. RhoA and Rac1 are differently expressed after the EZR silencing with respect to the PLCE silencing. The modulation of RhoA is described in the 143B cells proportionate to the PLC ε expression (142), suggesting that RhoA and PLC ε may concurrently participate in the same pathway.
In the Hs888 cell line, there are decreases in Ezrin, RhoA, and Rac1 simultaneous with an increase in PLC β1, suggesting the different and complicated regulation of Ezrin activity in Hs888 with respect to the other OS cell lines. Moreover, the EZR silencing in Hs888 seems to affect PLC genes, suggesting that Ezrin interacts with the same isoforms under normal conditions. The other studies addressing the crosstalk and the ordered timing of action of Ezrin and its networks in each OS cell line conclude that PLCs and Ras GTPases may contribute to highlighting the specific role of each molecule, providing new insights to the metastatic progression of the disease and new therapeutic strategies.
2.11. Role of OS Upon Microenvironment
Analyzing the role of factors produced by cells in the surrounding environment has recently offered promising insights in some research fields. The cell microenvironment encompasses molecules and factors, which can directly or indirectly act via biophysical, biochemical, or other routes and influence the cell metabolism and survival, growth, apoptosis, and proliferation, Upon nearby cells and distant immune cells (161). The detection of the nature and activity of the molecules involved in the microenvironment, including those belonging to signal transduction systems, might help to understand both physiological events and the systemic development of diseases (162) since additional/abnormal components may modify the morphological and functional features of cells (163).
In tumors, factors released in the microenvironment allow malignant cells to move from the primary tumor site, migrate, and establish a new site (162). Malignant cells can produce and release molecules in the microenvironment, and the immune system cells (e.g., cytokines, chemokines and growth factors) respond to such molecules. In vitro experiments have indicated that the modification of the components in the microenvironment interacts and affects the behaviour of both cells in the surrounding normal tissues and cells belonging to the immune system (162). Accordingly, the manipulation of the microenvironment may provide promising perspectives in terms of molecular therapy approaches. Immune system cells can behave differently depending on the signal transduction pathways acting in the microenvironment. In this regard, the OS supernatants have recently been used in vitro to modify the microenvironment (164).
The OS-derived supernatants are used to culture the THP-1 cells currently used as an experimental model in examining the monocytes/macrophage lineage differentiation (165). Supernatants from the cultured OS cell lines contain cytokines/chemokines and affect the cell survival and proliferation, induce differentiation, and modify the expression panel of the PLC enzymes (164). More specifically, the panel of cytokines differs depending on the OS cell line and differently affects the cell growth, proliferation, and survival.
The most remarkable effect on the growth rate of THP-1 cells is observed for the supernatant from the highly aggressive 143B cell line (115). The OS supernatants modify the morphological and functional features of the THP-1 cells, inducing differentiation into macrophages. This is probably due to the cytokines and chemokines released in the supernatants. In this regard, IL-2, IL-6, IL8, TNFα, and GM-CSF are detected in supernatants from MG-63, 143B, and Hs888 cell lines, and IL-1α and IL-1β are identified in the supernatants from 143B cell line (164). IL-2 is a central factor in controlling the immune response, and the remaining cytokines play pro-inflammatory roles (166). Both GM-CSF and TNF-α induce the differentiation of THP-1 into dendritic cells (165, 167).
Previous reports demonstrated that, the PLC enzymes are differently expressed in un-polarized M0, M1, and M2 macrophages (63). THP-1 cells express all PLC isoforms, with the exception of PLC (164). The supernatants from the OS cell lines reduce the expression of PLCs in THP-1 cells with particular attention to some isoforms, but with the remarkable exception of PLCB3 (164). The modification of PLCs’ expression slightly varies depending on the OS cells line, which in general decreases the transcription of selected isoforms.
The supernatant from the cultures of 143B cells reduces the expression of all PLC isoforms, including PLCD4 and PLCE that are not expressed. The supernatant from MG-63 cells block the transcription of PLCB1, PLCD4, PLCE, PLCH1, and PLCH2 and reduces the transcription of the remaining isoforms.
The supernatant from the cultures of HS888 cells blocks the transcription of PLCB1, PLCG2, PLCD4, PLCE, PLCH1, and PLCH2 and reduces the transcription of most of the other isoforms. The only exception is PLCB3, which is slightly increased. The supernatant from the cultures of MG-63 cells block the transcription of PLCB1, PLCD4, PLCE, PLCH1, and PLCH2, and the other isoforms are reduced. In this regard, the decrease is significant for PLCB1 and PLCG2.
PLCB1 and PLCG2 genes codify for isoforms belonging to primary PLCs, respectively PLC β1 and PLC γ2. PLC β1, involved in myocyte (133) and adipocyte differentiation (134), is correlated with Cyclin D3, which regulates signaling pathways acting in macrophages (168). The loss of PLC β1 heterozygosity acts upon the leukemic progression of myelodysplastic syndrome (42, 137). PLC β1 is also suggested to be involved in cell migration, as discussed in the next paragraph. PLC γ2 is highly expressed in cells belonging to the hematopoietic lineage and mainly acts in the immune system regulation (169, 170). Mutations in PLCG2 gene lead to auto-inflammatory diseases with immunodeficiency (171). PLC δ4 is expressed in response to mitogenic stimulation acting in cell growth and tumorigenesis, and PLC ε is involved in Ras and Rho signaling (172).
Accordingly, the block of transcription of PLC genes in the 143B cell line might suggest that OS supernatants are involved in the differentiation of THP-1 cells into macrophages. According to some researchers (63), these isoforms may represent the markers of macrophage differentiation. The PLCB3 gene in HS888 cells slightly increases and is the only one up-regulated isoform. The PLC β3 enzyme is expressed in some tissues (37) and is an upstream target of protein kinase C ε. PLC β3 is involved in the inflammatory response (173) and is a key to macrophage survival (174). PLCB3 silencing potentiates the Toll-like receptor’s inflammatory signaling cascade in cystic fibrosis bronchial epithelial cells, also inducing IL-8 release (175).
To sum up, the OS supernatants mimic the variations that in vivo OS may drive in the microenvironment. Most likely, OS supernatants contain further factors other than the aforementioned cytokines. The effects of OS supernatants on the proliferation and differentiation of THP-1 and the expression of PLC enzymes suggest that tumors can modify the microenvironment composition. The effects of the OS supernatants on the THP-1 cells confirm that modifications in the microenvironment can affect the immune system cells and probably interfere with the progression of the diseases.
2.12. Extracellular Vesicles from OS
The role of extracellular vesicles (EVs), a recently identified mechanism of communication among cells, has provided promising insights into the research. EVs are particles coated by a membrane lipid-bilayer produced by the cell and released in the microenvironment (176). The lumen of EVs, surrounded by its own membrane, contains molecules such as DNA, RNA, miRNA, and proteins, derived and transferred out from the cell (177). Thanks to EVs’ transfer, a cell can act upon other cells located in the surrounding tissue and/or in distant target organs. Furthermore, malignant cells can share membrane molecules, intracellular proteins, and genetic information, which might underlie events leading to metastasis formation (178). More recently, research has addressed the proteins/phospholipids in the EVs’ membrane, and PLC enzymes are described in the EVs produced by human osteoblasts and OS cell lines (179).
Osteoblasts and OS cell lines release PLCs-containing EVs at different levels, suggesting that EVs in the bone are used by the cell to vehicle PLC enzymes and transduce signals (179). The expression panel of PLCs and their localization inside the EVs slightly differ depending on the specific cell lines.
All PLC isoforms are detected in EVs from osteoblasts and OS cells; however, they are differently localized in the vesicle compartments. As PLC enzymes work at the cell membrane and EVs are vesicle coated with cell membrane, we expected to find them in the EVs. Some PLCs are described both in the EV’s membrane and as cargo, others in the membrane or within the vesicle (179).
More evidently, PLC β1 is conveyed by EVs in OS cells compared to control osteoblasts, suggesting a role in EV-mediated effects. In addition to the aforementioned roles of PLC β1 and its role in metastatic cell migration through lipid-dependent sequestration of cofilin (180), both PLC β1 and PLC η2 are described in HUVEC podosome-like structures, suggesting their involvement in cytoskeleton remodeling and subsequent cell migration (66).
The PLC γ sub-family is probably involved in the early steps of cell migration. In addition to the aforementioned roles, PLC γ isoforms induce actin polymerization leading to the formation of cell protrusions, involved in sticking to the extracellular matrix, controlling migration direction, and initiating cell crawling (181). PLC γ2 was identified in the EV’s membrane, more markedly in the 143B cell line (179), corroborating the previous hypothesis indicating that PLC γ2 plays a role in aggressiveness (115, 142).
The presence of PLC enzymes in EVs suggests that Osteosarcoma can affect both the surrounding microenvironment and distant target cells, providing the niche for metastasis. Further studies are recommended to investigate the role of EV-mediated PLC signaling, both in the microenvironment and in reservoir cells, to provide insights into issues leading to the OS spreading. Different levels and differently-located PLCs containing EVs are produced by osteoblasts and each specific OS cell line, opening promising perspectives. Futures researchers are suggested to address this issue in detail. In this regard, EVs collected from blood and the content of their PLCs might be used as diagnostic biomarkers for OS and might be used as a prognostic feature to monitor progress.
3. Conclusions
Metastasis spreading aggravates the prognosis of the clinical outcomes in patients affected with OS. Despite many research efforts and recent improvements, the identification of factors and molecules involved in the metastatic spreading of the OS tumors is largely unknown. Accordingly, detecting the origin, behavior, interconnections, and interactions of molecules involved in the formation of metastases may contribute to understanding the tumor dissemination mechanisms, opening the way to novel therapeutic strategies. The prominent role of Ezrin and its relationship with both the PI signal transduction pathway and the complex Ras GTPase system provide new insights into issues leading to metastasis formation in OS, opening promising perspectives to refine the prognosis and develop new targeted molecules for therapy.