1. Context
Colorectal cancer (CRC) is one of the most common neoplasms and leading causes of cancer-specific mortality worldwide (1, 2). Major treatment modalities in rectal cancer are surgery, radiation, and chemotherapy. Individuals with recurrent or metastatic disease may salvage via surgery; however, chemotherapy remains the dominant therapy in many cases (3-5).
A major challenge in CRC treatment is drug resistance that is directly associated with low survival rates. A better understanding of acquired and intrinsic therapeutic resistance will play a principal role in the effective development of treatment strategies (6, 7). Although resistance to treatment is quite common, more comprehensive molecular mechanisms involved in chemical resistance and characterizing their effects reveal new targets to improve tumor response and facilitate the development of effective treatment programs to overcome resistance (8, 9). Recently, discovering new cancer biomarkers has gained much attention in cancer investigations. Cancer biomarkers are considered detectable alterations in any molecular marker at the metabolite, protein, DNA, and RNA level. Non-coding RNAs as a group of transcribed functional RNA molecules lack protein-coding capacity (10, 11).
Non-coding RNAs consist of long non-coding RNAs (lncRNAs) and small RNA. The small RNA typically ranges from 20 to 30 nt and includes miRNAs, piRNAs, and endogenous siRNAs, while lncRNAs usually exceed 200 nucleotides in length (12). lncRNAs can control the expression level of genes by a different mechanism such as microRNA sponge, RNA decoy, translation inhibition, and chromatin modification (13, 14). Moreover, lncRNAs contribute to regulating biological processes such as differentiation, apoptosis, proliferation, migration, and invasion (15, 16). MiRNAs can decrease the expression level of translated protein through the interaction of miRNAs with target mRNA. Furthermore, histone modifications, DNA methylation, and RNA modification can regulate the expression of miRNAs (17-19). Due to stability and presence in biological fluids, lncRNAs and miRNAs can be used as a non-invasive diagnosis, prognosis, and response to treatment biomarkers for CRC (20, 21). Considering the importance of miRNAs and lncRNAs as prognostic and response to treatment biomarkers, in this review, we discuss the role of lncRNAs and miRNAs in CRC chemoresistance.
2. Role of MicroRNAs in CRC Chemoresistance
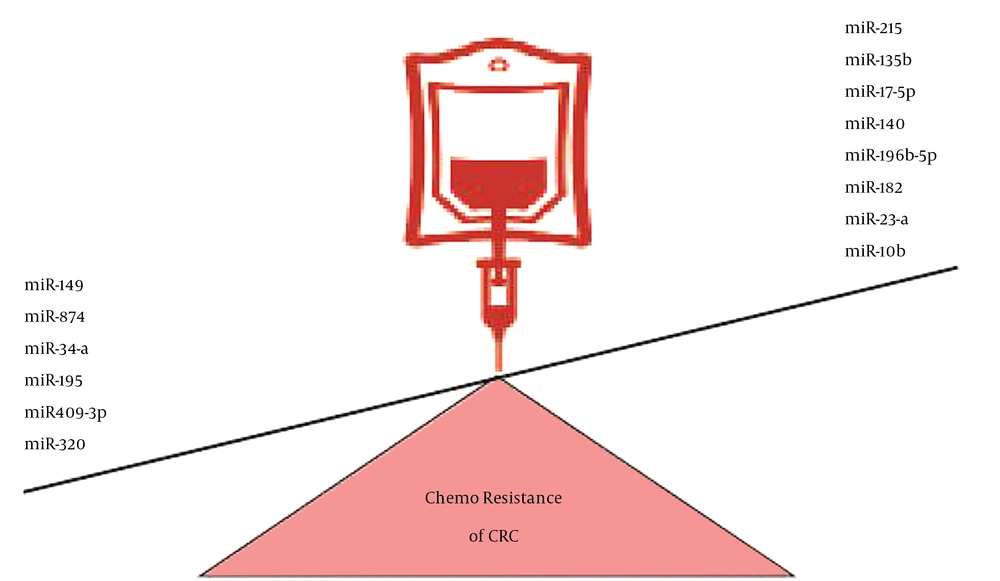
Numerous miRNAs, including miR-203, miR-140, miR-17-5p, miR-215, miR-149, miR-135b, miR-182, miR-23a, miR-874, miR-34a, miR-10b, miR-196b-5p, miR-195, miR-409-3p, and miR-320, have a regulatory role in the response of colorectal tumor cells to therapeutic agents (Figure 1) (4, 22-36).
Various miRNAs with a regulatory role in the response of CRC cells to therapeutic agents; the downregulation of miR149, miR874, miR34-a, miR195, miR409-3p, and miR320 and the upregulation of miR17-5p, miR215, miR135b, miR140, miR196b-5p, miR182, miR23-a, and miR10b were associated with CRC chemoresistance.
MiR-203 has an essential role in regulating proliferation, migration, and invasion of CRC. This miRNA can be used as a prognosis and diagnosis biomarker (37, 38). Li et al. indicated that the upregulation of miR-203 led to decreasing proliferation but increasing apoptosis and chemosensitivity to paclitaxel in p53-mutated colon cancer cells. On the other hand, the chemosensitizing effects of miR-203 are mediated by Akt2 inhibition and subsequently MTDH and HSP90 downregulation. Furthermore, active caspase-3, the Bax/Bcl-xL ratio, and apoptosis increase due to miR-203 upregulation (22). It has also been shown by Zhou et al. that the expression level of miR-203 was correlated with resistance to oxaliplatin in CRC cells. The downregulation of miR-203 leads to oxaliplatin chemosensitization by targeting the ataxia telangiectasia mutated (ATM) (23).
MiR-140, miR-17, and miR-215 have different effects on CRC. MiR‑140‑5p and miR-215 as tumor suppressor miRNAs suppress the invasion and proliferation of CRC. MiR-17 endorses the invasion and proliferation of CRC (39-42). Song et al. showed that miR-140 was upregulated in CRC stem cells. On the other hand, knocking down miR-140 leads to 5-FU chemosensitization in CRC by targeting the Histone deacetylase 4 (HDAC4) (24). Fang et al. indicated that the upregulation of miR-17-5p led to distant metastasis and low survival in CRC patients. Moreover, the expression level of miR-17-5p is associated with multiple drug-resistance by targeting PTEN (25). Song et al. pointed out that miR-215 led to the resistance of colon cancer cells to Tomudex (TDX) and methotrexate (MTX) by targeting dihydrofolate reductase (DHFR) or thymidylate synthase (TS) (26).
miR-149, miR-135b, and miR-135b regulate the migration and invasion of CRC (43-45). Liu et al. showed that the expression level of miR-149 was associated with the chemosensitivity of colorectal tumor cells to 5-FU and that the upregulation of miR-149 led to apoptosis induced by 5-FU via targeting the forkhead box transcription factor (FOXM1) (27). He et al. reported that the overexpression of miR-135b reduced the chemosensitivity of CRC cells to doxorubicin and apoptosis induced by doxorubicin through targeting the large tumor suppressor kinase 2 (LATS2) (28). Also, Liu et al. showed that the upregulation of miR-182 and MiR-135b was associated with the chemoresistance of CRC cells to 5-FU by targeting ST6GALNAC2 (29).
miR-23a and miR-196b-5p endorse the invasion, migration, and metastasis of CRC (46, 47). On the other hand, miR-195 and miR-10 have regulatory effects on the proliferation and migration of CRC cells (31, 48-50). Li et al. showed that miR-23a attenuated the 5-FU induced apoptosis and led to the chemoresistance of microsatellite instability CRC by targeting ABCF1 (51). Han et al. found that miR-874 led to apoptosis induction by 5-FU and chemosensitivity to 5-FU through the X-linked inhibitor of apoptosis protein (XIAP) targeting (31). Li et al. reported that the upregulation of mir-34a led to the chemosensitivity of CRC to 5-FU through LDHA targeting (32). Recent studies have indicated that expression levels of miR-10b and miR-196b-5p are correlated with poor prognosis in CRC patients. miR-10b enhances the 5-FU resistance of CRC cells by targeting BIM. miR-196b enhances 5-FU resistance and apoptosis induction by 5-FU by targeting SOCS3 and SOCS1 (4, 33). Qu et al. showed that a low expression level of miR-195 was associated with resistance to doxorubicin in CRC cells. Moreover, the overexpression of mir-195 promotes the chemosensitivity of CRC cells to doxorubicin and doxorubicin-induced apoptosis by targeting BCL2L2 (34).
In addition, miR-409-3p can regulate the invasion and metastasis of CRC (36, 52). Tan et al. showed that the downregulation of miR-409-3p was associated with the oxaliplatin resistance of CRC cells. Furthermore, the upregulation of miR-409-3p leads to the chemosensitivity of CRC cells to oxaliplatin by targeting Beclin-1, which regulates autophagy (35). On the other hand, Wan et al. found that miR-320 enhanced the chemosensitivity of CRC cells to 5-FU and Oxaliplatin by targeting FOXM1 and inhibiting the Wnt/b-catenin pathway (36).
3. Role of lncRNA in CRC Chemoresistance
Numerous IncRNAs, including H19, SCARNA2, PVT1, UCA1, MEG3, ANRIL, BANCR, HOTAIR, MALAT1, XIST, ENST00000547547, CRNDE, SLC25A25-AS1, GIHCG, CASC15, TUG1, ENST00000547547, and LINC00152 have a regulatory role in the response of colorectal tumor cells to therapeutic agents (Table 1) (53-69).
| lncRNA in Chemoresistance | Mechanism | Expression | References |
|---|---|---|---|
| H19 | SIRT1 mediated autophagy | Overexpressed | (53) |
| SCARNA2 | Targeting the miR‐342‐3p and affecting the EGFR/BCL2 pathway | Overexpressed | (54, 70) |
| PVT1 | Apoptosis inhibition and regulating the Bcl-2, MRP1 mTOR, and P-gp | Overexpressed | (70) |
| UCA1 | Influencing apoptosis and proliferation | Overexpressed | (56) |
| MEG3 | Targeting the miR-141 and affecting the PDCD4 expression level; Increasing the chemotherapy-induced apoptosis | Decreased | (57, 71) |
| ANRIL | Targeting Let-7a and affecting the ATP-binding cassette subfamily C member 1 Influencing apoptosis and proliferation | Overexpressed | (58) |
| BANCR | Regulating the CSE1L expression level through sponging miR-203 | Overexpressed | (59) |
| HOTAIR | Targeting miR-203a-3p and miR-218 and affecting the Wnt/β-catenin and NF-κB signaling pathway | Overexpressed | (60, 72) |
| MALAT1 | Controlling the chemotherapy-induced EMT, and regulating E-cadherin expression | Overexpressed | (73) |
| XIST | Targeting miR-124 and affecting SGK1 | Overexpressed | (62) |
| CRNDE | Targeting miR-181a and regulating the Wnt/β-catenin signaling pathway | Overexpressed | (64) |
| CASC15 | Targeting miR-145 and regulating ABCC1. | Overexpressed | (67) |
| LINC00152 | Regulating chemotherapy-induced apoptosis and controlling the NOTCH1 expression level through sponging miR-139-5p | Overexpressed | (69) |
| ENST00000547547 | Targeting miR-31 | Decreased | (63) |
| TUG1 | Regulating CPEB2 through sponging miR-186 | Overexpressed | (68) |
Recent studies have indicated that H19, MEG3, LINC01296, HOTAIR, and SCARNA2 are related to the prognosis of CRC patients (54, 71, 74-76).
Recent studies have also shown that H19 leads to CRC chemoresistance to 5-FU through SIRT1 mediated autophagy (53, 77). Zhang et al. demonstrated that CRC chemoresistance to oxaliplatin or 5‐FU correlated with SCARNA2 expression. On the other hand, SCARNA2 promotes chemoresistance by targeting the miR‐342‐3p‐EGFR/BCL2 pathway (54). Zhu et al. discovered that XIST was upregulated in the doxorubicin-resistant CRC tissue, whereas miR-124 expression showed downregulation. By XIST knockdown, the doxorubicin resistance is hindered in CRC cells (62). Experimental data demonstrated that the downregulation of MEG3 was apparent in oxaliplatin-resistant CRC cells. However, the upregulation of MEG3 enhances the sensitivity of chemoresistance cells to oxaliplatin (57, 71). Liu et al. reported that LINC01296 promotes 5-FU resistance, cell proliferation, and metastasis through the miR-26a/GALNT3 axis (78). In a study by Xiao et al., HOTAIR could regulate the chemoresistance and progression of CRC through the modulation of miR-203a-3p expression and the activity of the Wnt/β-catenin pathway (72). It is also reported that HOTAIR is involved in 5-FU resistance in CRC by inhibiting the miR-218 pathway and activating the NF-κB pathway (60).
ANRIL, UCA1, MALAT1, ENST00000547547, CRNDE, GIHCG, BANCR, TUG1, ENST00000547547, LINC00152, and CASC15 have a role in the regulation of proliferation and metastasis of CRC (59, 66, 69, 79-85). Recently, in cetuximab-resistant cancerous cells, UCA1 expression has been shown to be significantly high. UCA1 can decrease apoptosis but increase cell proliferation and 5-FU resistance through miR-204-5p suppression in CRC (56). Another research indicated that the overexpression of ANRIL led to the oxaliplatin and 5-FU resistance of CRC through the Let-7a/ABCC1 axis (58). The discovery of the MALAT1 lncRNA as an inducer of oxymatrine resistance in CRC may produce prognostic and therapeutic information for patients (61). The knockdown of MALAT1 increases the expression level of E-cadherin and inhibits oxaliplatin-induced EMT. Moreover, MALAT1 interaction with miR-218 shows their essential effect on the prognosis of patients who are under treatment of standard FOLFOX. These findings indicate how MALAT1 presents a chemoresistant function in CRC (86). Evidence suggests that ENST00000547547 overexpression decreases the chemoresistance of 5-FU and increases the 5-FU induced cell apoptosis of CRC by competitive binding to miR-31 (63). Gao et al. demonstrated that CRNDE promoted oxaliplatin resistance and tumor metastasis by miR-136 inhibition in CRC (87). Also, Han et al. characterized miR-181a-5p as a target of CRNDE. Knocking down CRNDE leads to the chemosensitivity of CRC through the MiR-181a-5p/Wnt/β-catenin axis (64). Jiang et al. showed that 5-FU and oxaliplatin-resistant CRC cells gain excessive expression levels of GIHCG (66). Ma et al. indicated that the expression level of BANCR was associated with tumorigenesis and adriamycin resistance via controlling the miR-203/CSE1L axis (58, 59). Another study demonstrated that TUG1 lncRNA mediated methotrexate resistance in CRC by regulating CPEB2 through binding miR-186 (68). Li et al. showed that ENST00000547547 through competitive binding with miR-31 reduced 5-FU-resistant CRC (63). Another study discovered that LINC00152 was overexpressed in the CRC tissue and negatively correlated with CRC patients' survival rates. LINC00152 could associate with the 5-FU resistance of CRC through the miR-139-5p/NOTCH1 axis (69). Gao et al. indicated the increased expression level of CASC15 oxaliplatin-resistant CRC. CASC15 promotes the oxaliplatin resistance of CRC through the miR-145/ABCC1 axis (67).
PVT1 has a role in regulating the proliferation and apoptosis of colorectal tumor cells (88). The up-regulation of XIST and the down-regulation of SLC25A25-AS1 promote colorectal tumor cell proliferation (65, 89). Ping et al. suggested that the excessive expression of PVT1 considerably increased cisplatin resistance to colorectal tumor cells (55). Moreover, PVT1 is exceedingly upregulated in 5-FU resistant CRC patients and cells. Thus, PVT1, with its important regulatory effect on tumorigenesis and chemoresistance, can be introduced as a significant target in CRC treatment (70). The 5-FU resistance can be reversed by the knockdown of XIST, whereas increased levels of XIST can limit cytotoxic effects of 5-FU (90). Zhu et al. discovered that the expression level of XIST increased in the doxorubicin-resistant CRC tissue, whereas miR-124 was downregulated. By XIST knockdown, doxorubicin resistance was hindered in CRC cells (62). Recent studies have revealed that the downregulation of SLC25A25-AS1 increases EMT and chemoresistance and that its excessive expression prevents the proliferation of CRC (65).
4. Conclusions
Despite many improvements in chemotherapy, a significant challenge for this modality is resistance to anticancer drugs. Due to its substantial clinical significance, chemoresistance has led scientists to identify pathways and mechanisms involved in resistance to chemotherapy. Many large-scale molecular studies have been performed to investigate the role of lncRNAs and microRNAs in tumor progression, metastasis, and response to tumor treatment. This study shows that lncRNAs and microRNAs play diverse roles in CRC chemoresistance. Thus, non-coding RNA can be used as a potential response to treatment biomarker for CRC.