1. Background
One of the most significant achievements of medical sciences is beta-lactams. In this regard, the emergence of resistance to beta-lactam antibiotics has become a global concern. The coding gene of this enzyme is abundant in the E. coli strains. This made the World Health Organization (WHO) to highlight the risk of developing antibiotic-resistant strains. The WHO recommended the WHONET software to health centers to inform physicians about the results of local and global antibiotic resistance (1).
Phenotypic tests are the simplest techniques for the detection of antibiotic resistance worldwide. However, the phenotypical tests fail to detect the extended-spectrum beta-lactamase (ESBL) antibiotic resistance fully (2-5).
The Multiplex PCP method can detect 2 or 3 target sequences simultaneously. The rapid and exact detection and reports on the findings can help physicians treat their patients. Beta-lactam antibiotics are antibiotics effective in treating bacterial infections worldwide (2, 3, 5).
Since beta-lactams are inactive by β-lactamases, there is a need to use appropriate antibiotics to prevent the spread of antibiotic resistance genes.
ESBLs are enzymes inactivating beta-lactam antibiotics by attacking to beta-lactam ring (2-5).
ESBL refers to a group of enzymes hydrolyzing beta-lactam antibiotics such as penicillin, cephalosporin, cephamycin, and monobactam. The most abundant type of ESBL is the CTX-M beta-lactamase (6-9).
The widespread use of antibiotics and the advent of drug resistance concerning the prevalence of ESBLs in the strains are serious concerns. Beta-lactam is one of the antibiotics used against these bacteria. In recent years, the occurrence of multi-drug-resistance (MDR) isolates has aroused many concerns about selecting the appropriate antibiotic pattern. Accordingly, molecular techniques for the rapid and correct identification of antibiotic resistance are valuable (10-14).
2. Objectives
This study aimed to set up and design a Multiplex PCR technique for detecting extended beta-lactamase genes using the Multiplex PCR method.
3. Methods
3.1. Sample Collection
Several 50 E. coli clinical isolates were collected from the laboratory of Baqiyatallah Hospital and were confirmed by biochemical tests. Disc combine was used for the ESBL phenotypic identification.
3.2. Design Primer
First, complete DNA sequences were searched on the NCBI. The primers were designed with the Genscript software. The forward and reverse primers were blasted, and the results confirmed the suitability of the designed primers. Three primer pairs were evaluated with Oligo Analyzer software. Then the primers were evaluated with silico PCR amplification software (Table 1).
| Gene | Forward-Primer | Reverse-Primer | Tm | Amplicon Size (bp) |
|---|---|---|---|---|
| TEM | GAGGACCGAAGGAGCTAACC | TTGCCGGGAAGCTAGAGTAA | 60 | 188 |
| AmpC | CTCGACCTCGCGACCTATAC | CTGCCACTGGCGGTAGTAGT | 60 | 102 |
| KPC | CAGCTCATTCAAGGGCTTTC | GTCCAGACGGAACGTGGTAT | 60 | 283 |
3.3. Determining Primer Sensitivity
Primer sensitivity was studied with different DNA dilutions. The nucleic acid serial dilution was prepared, and for all genomic dilutions, PCR was performed. The lowest dilution of the PCR reaction was set as test sensitivity.
3.4. Determining Primer Specificity
To obtain the specificity of the primers, the PCR reaction was performed considering the above conditions for the nucleic acid Staphylococcus aureus and Bacillus subtilis.
3.5.DNA Extraction
Nucleic acid extraction was carried out by the Cinnaclon kit. The purity and concentration of the nucleic acid were measured by spectrophotometry Nanodrop.
3.6.PCR Reaction
The PCR was carried out to amplify the genes, and the final volume of the PCR reaction was 25 µL. (Tables 2 and 3).
| Components | Concentration |
|---|---|
| Master mix (1x) | 12.5 µL (1x) |
| Forward primer (0.1 - 1 µm) | 1 µL (10 µm) |
| Reverse primer (0.1 - 1 µm) | 1 µL (10 µm) |
| Template DNA | 1 µL (20 pg) |
| Sterile deionized water | 9.5 µL |
| Total volume | 25 µL |
| Cycle PCR | Temperature | Time (Second) | Number Cycle |
|---|---|---|---|
| Primary denaturation | 95°C | 300 | 1 |
| Second denaturation | 95°C | 30 | 35 |
| Annealing | 60°C | 45 | 35 |
| Extension | 72°C | 40 | 35 |
| Final extension | 72°C | 300 | 1 |
3.7. Gel Electrophoresis
The DNA amplified by PCR was observed on a 1% (w/v) agarose gel electrophoresis with SYBR® Safe (Qiagen) color.
4. Results
4.1. Antibiogram Test
Disc diffusion was evaluated in 50 bacterial samples isolated in the Baqiyatallah Hospital (Table 4).
| Antibiotics | Resistant (%) | Intermediate (%) | Sensitive (%) |
|---|---|---|---|
| Ofloxacin (OFX) | 96 | 0 | 4 |
| Amikacin (AN) | 45 | 30 | 25 |
| Piperacillin (PIP) | 98 | 1 | 1 |
| Ciprofloxacin (CP) | 96 | 2 | 2 |
| Gentamicin (GM) | 79 | 6 | 15 |
| Cefotaxime (CTX) | 98 | 1 | 1 |
| Nitrofurantoin (FM) | 49 | 14 | 35 |
| Imipenem (IMI) | 39 | 11 | 50 |
| Meropenem (MER) | 30 | 5 | 65 |
| Norfloxacin (NOR) | 96 | 3 | 1 |
4.2. Multiplex PCR Results
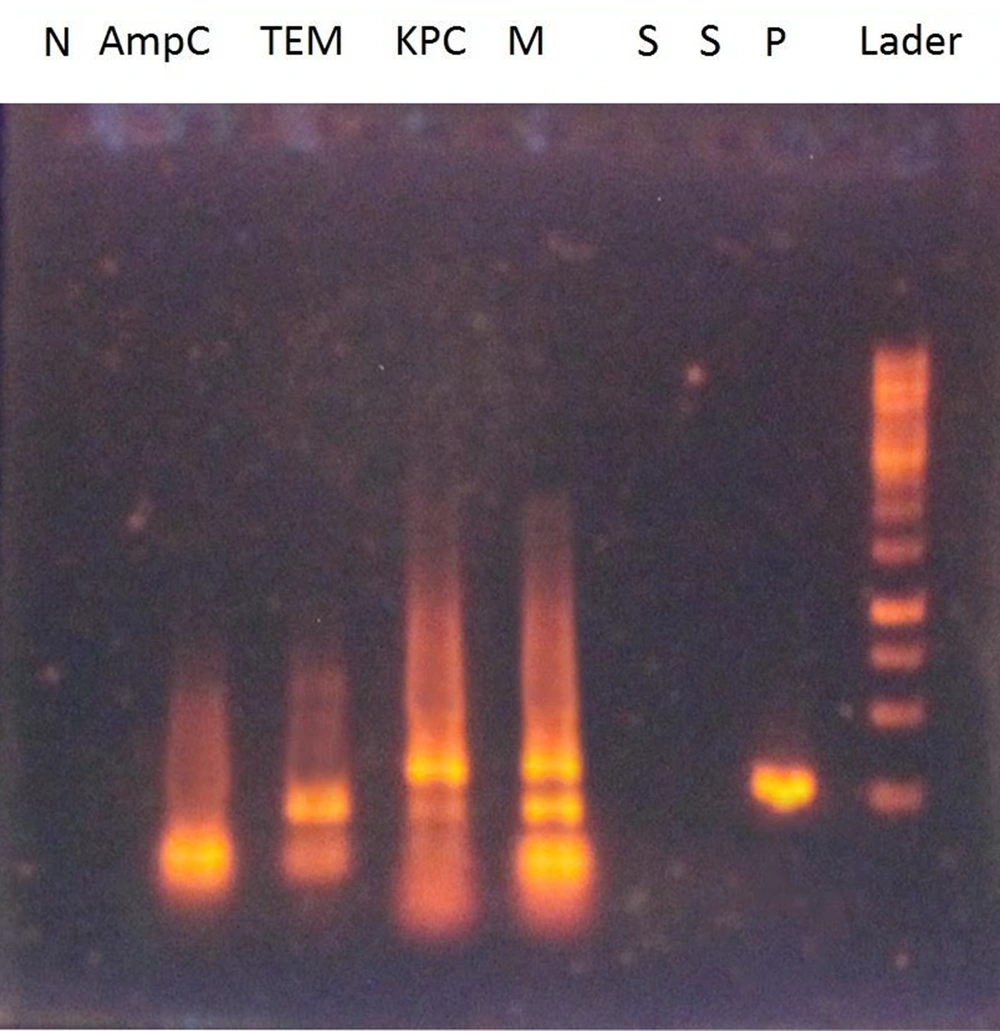
The multiplex PCR results for the ESBLs of AmpC, TEM, and KPC on 1% agarose gel are illustrated in Figure 1. The sensitivity and specificity of the multiplex PCR technique for ESBL genes were 0.001 ng and 100%, respectively.
Multiplex PCR results for ESBL genes on gel agarose 1%. S: Wells for primer specificity containing primers with DNA S. aureus and B. subtilis; N: Wells containing primers without DNA as a negative control; P: Wells containing primers and DNA containing ESBL gene as a positive clinical control; M: Well-containing three-pair primers of ESBL genes; AmpC: Well containing AmpC gene primer; TEM: Well containing TEM gene primer; KPC: Well containing KPC gene primer.
4.3. Frequency of ESBL Genes
The confirmed primers of TEM, KPC, and AmpC genes were evaluated in the 50 Escherichia coli clinical isolates (Table 5).
| Genes | Resistant, Frequency % |
|---|---|
| TEM | 58 |
| KPC | 18 |
| AmpC | 84 |
5. Discussion
The extensive use of antibiotics affected the prevalence of antibiotic resistance genes in different regions. The increased prescription of broad-bet-lactams and long hospitalization caused the spread of antibiotic-resistant genes.
Beta-lactams are one of the most common antimicrobial agents in treating serious infections induced by Enterobacteriaceae infections. The emergence of resistance to beta-lactams antibiotics have resulted in many treatment failures (10, 15-19).
The present study used the multiplex PCR technique to detect the ESBL TEM, KPC, and AmpC genes simultaneously.
The sensitivity of the multiplex PCR for the TEM, KPC, and AmpC genes was 0.001 ng, and the test specificity was 100%. The results indicated that molecular and phenotypic tests were compatible.
This method decreases the use of costly and widespread antibiotics and promotes preparation for the rapid diagnosis of antibiotic resistance when epidemics occur. Given the significance of identifying multi-drug resistance bacteria, it is necessary to design appropriate methods for the rapid and correct detection of antibiotic resistance (10, 15-19).
The resistance to these antibiotics is caused by genetic structures such as integrons, transposons, and plasmids; therefore, there is concerns about the transmission of antibiotic resistance genes to other bacteria (17, 18).
In the present study, resistance to Cefotaxime and piperacillin was reported; however, the isolates of pandemic-drug-resistance (PDR) were not reported. The multi-drug-resistance (MDR) and extensive drug-resistance (XDR) isolates were also reported (18, 19).
The present study showed that about 100% of E. coli isolates had an MDR pattern, which was of a great concern.
According to the present findings, the highest frequency of resistance to piperacillin and cefotaxime antibiotics was 98%. The highest rates of sensitivity to the Meropenem and Imipenem antibiotics were reported to be 55% and 50%, respectively. In his study in the Jahrom Hospital, Emamghorashi and Kohanteb reported antibiotic resistance to vancomycin, gentamicin, Nitrofurantoin, amikacin, and ciprofloxacin to be 56.2, 72.1, 34.3, 28.6, and 6.7%, respectively. In this study, the frequency of resistance to these antibiotics was significantly higher than in Emamghorashi and Kohanteb’s study. The acquisition and spread of antibiotic-resistance genes over time cause such inconsistency (20).
In a study by Mobasherizadeh et al., extent-spectrum β-lactamases from UTI infections in admitted and outpatient patients were isolated in Esfahan, and the resistance rates of Klebsiella pneumonia and Escherichia coli producing β-lactamases were 41.6 and 47.97%, respectively, suggesting the lowest and highest antibiotic resistance for nitrofurantoin (16.7%) and cotrimoxazole (75%), respectively (21).
Kaikha and Rava in Zahedan reported the antibiotic resistance rates of E. coli to the antibiotics nitrofurantoin, amikacin, ceftazidime, and gentamicin were 26.1, 19.5, 44.8, 13.7, and 4.5%, respectively. Their reported values were below the ones reported in the present study (22).
In Heidari-Soureshjani et al.’s study, the rates of antibiotic resistance to nalidixic acid, cotrimoxazole, nitrofurantoin, ciprofloxacin, cefotaxime, gentamicin, and imipenem were 67, 21, 32, 8, 49, 7, 43, and 38%, respectively (23).
The ESBL genes are effective agents in emerging resistance to beta-lactam antibiotics. Organisms containing ESBL genes are more pathogenic and virulent; hence, the timely and accurate diagnosis of this type of resistance, reporting the findings to physicians, and the provision of an appropriate medical advice can reduce patient problems and promote their treatment. In this study, fifty-eight percent of the isolates contained the TEM gene, eighteen percent of the isolates contained the KPC gene, and eighty-four percent of the isolates contained the AmpC gene. Furthermore, in the present study, 58% of the samples contained the KPC, TEM, and AmpC genes simultaneously. Shahcheraghi et al.'s study showed that 24% of isolates contained the TEM gene. This frequency is lower than the value reported in the present study. The distribution of the resistant plasmids among E.coli isolates probably arouses antibiotic resistance (24). In 2008, a comprehensive study on ESBL enzymes was conducted in Switzerland, which showed that 42.9% of the isolates contained the AmpC gene.
The studies that used the PCR method for detecting ESBL genes showed that nearly 50% of reported isolates contain TEM genes, which was compatible with the findings of the present study (1, 25-32). Recent studies have indicated that resistance to beta-lactam induced by ESBL is increasing rapidly. Possible reasons are the improper administration of antibiotics, the lack of appropriate methods in identifying antibiotic resistance, and the improper interpretation of new identification methods.
5.1. Conclusions
Antibiotic resistance, the prevalence of broad-spectrum beta-lactamase genes, and the extent of spectrum-beta-lactamase in gram-negative bacteria are global concerns and require infection control management. Regarding the immediate and accurate detection of these strains, the multiplex method proposed in this study would help physicians to prescribe appropriate antibiotics and avoid extended hospital stays and high costs.