1. Background
One of the first photosynthetic prokaryotes on Earth is cyanobacteria, and the oxygen released into the atmosphere by their photosynthesis is an important precursor to the ozone layer (1). Their successful adaptation to life in various ecological niches is mainly attributed to genetic diversity and the production of important natural products (1). Natural cyanobacteria products are also a potential source of drugs for developing therapeutic agents used to treat diseases such as cancer, parasitic infections, and inflammation. The primary sources of these natural compounds are Oscillatoriales, Nostocales, Chroococcales, and Synechococcales. More than 550 secondary metabolites have been reported in different types of cyanobacteria, including Oscillatoria and Lyngbya (2). Most of these compounds contain nitrogen and are produced from modular biosynthetic enzymes such as nonribosomal peptide synthetases (NRPS), polyketide synthase (PKS) and their combinations. In addition, most of the secondary cyanobacterial metabolites are peptides and depsipeptides. Most of these compounds are synthesized by NRPS or hybrid NRPS/PKS pathways, which are nonribosomal. These compounds include non-protein and methylated amino acids, which is characteristic of peptides that are biosynthesized by nonribosomal peptide synthetases (3).
Among the products obtained by these enzymes are antibiotics like penicillin and vancomycin (4). The multiple structural features in secondary metabolites and the production of these products, even in the presence of ribosomal inhibitors or RNase, are evidence of this assertion (5). nonribosomal peptide synthetases (NRPSs) belongs to the family of mega synthetases, known as the largest multimodular enzymes with a molecular weight of over 2 or 3 megadaltons consisting of repeated units called modules, each of which is approximately 1000 to 1500 long amino acids. Adding residues in the growing peptide chain is the function of a module. A complete module has at least three enzyme domains, including the adenylate domain (A), the condensation domain (C), and the thioled domain (T). Each module catalyzes a complete chain elongation cycle, including three basic reactions: precursor recognition, activation as acyl adenylate, and covalent binding as thioester. These enzyme activities are integrated as a distinct catalytic domain into each module. Adenylated domain controls the early stages of nonribosomal peptide synthesis and selects amino acid precursors and actives ingredient as aminoacyl adenylate. The unstable amino acid adenylate is transferred in succession downstream of the T-domain. Therefore, the T domain is also called peptidyl carrier protein (PCP). The condensation domain catalyzes the formation of a peptide bond between an aminoacyl PCP or peptidyl-PCP from the upstream module and an aminoacyl residue attached to the corresponding downstream module. The thioesterase or terminal domain is critical for product release and catalyzes the release of the biosynthetic peptide in linear, cyclic, or branched form.
These enzymes synthesize many biologically active natural peptides (6). Polyketide synthase (PKS) enzymes activate the amino acid substrate by consuming ATP in the form of aminoacyl adenylate. Polyketides are another critical group of natural products. Multimodular polyketide synthases synthesize a large number of them. Despite the high structural diversity of these products obtained by the reduction of keto groups through hydroxylation and glycosylation reactions, only two building blocks, acetate and propionate, are used to synthesize polyketides. The building blocks in activated forms are malonyl and methyl malonyl coenzyme A. How these enzymes produce the product is a little complex and is as follows. Each polyketide synthase module is joined to essential and, in some cases, optional domains for elongation. The three essential domains include the ketosynthase domain (KS), acyltransferase domain (AT), and thioesterase domain (Te). In the first step, polyketide aggregation is catalyzed by the acyltransferase domain (about 350 residues), which transfers the activated building blocks to the ACP domain. The ketosynthase domain (about 450 residues) catalyzes the elongation reaction in the synthesis of polyketides. The reaction starts with the transfer of the ketide chain from an upstream module's acyl carrier protein to the ketosynthase domain's active site. Then, a malonyl unit attached to the acyl carrier protein is decarboxylated by the ketosynthase domain. Finally, the thiolated domain releases the product at the C terminus (3).
Research on cyanobacteria in the pharmaceutical industry began in the late 1970s, resulting in the discovery of new compounds with antiviral, antifungal, anticancer, anti-HIV, antibacterial, and cytotoxic properties (7). The widespread use of antibiotics to treat humans and animals for over half a century has caused a dramatic decrease in the efficacy of many antibiotics due to the evolution of bacterial resistance. In addition, the number of new antibiotics introduced into the world has been significantly reduced, so searching for metabolites with new antibiotic properties is especially important (8, 9).
2. Objectives
Despite numerous studies on the distribution of the NRPS and PKS genes, none has examined the aquatic cyanobacteria strains of Tehran Cascade (8). Therefore, research to discover the presence of PKS and NRPS genes with the ability to synthesize antimicrobial compounds in 12 strains of fresh water is one of the first investigations to introduce cyanobacteria strains into the production of metabolites of medicinal origin. Hence, research to reveal the presence of polyketide synthase genes and nonribosomal peptide synthetases and the relationship between these genes and antimicrobial activities is significant.
3. Methods
3.1. Sampling and Cultivation of Cyanobacteria Samples
Water samples were collected from the surface to a depth of 10 cm from the Tehran waterfall with plastic bottles. For this purpose, the collection containers were carefully and slowly rotated to fill with water, mud, and algae. The sampler took care to leave the upper part of the containers empty to have enough air (10, 11). The BG-110 stock (Merck) solid culture medium (without nitrogen salts) was used to cultivate and purify cyanobacteria of Nostocales order. Then, the Petri dishes containing the inoculum of cyanobacteria were placed inside the growth chamber with a light intensity of 40 to 60 µmol/m2/s and a temperature of 28°C to 30°C (12). After two weeks, many cyanobacteria colonies were seen in each Petri dish.
3.2. Purification and Preparation of Mono-algal Culture for Molecular Taxonomic Investigation
The presence of movable filaments or hormogonium that could glide on the Petri dish is one of the characteristics of the Nostocales order, which was used to isolate and purify this order (12, 13). Finally, axenic samples were tested by inoculation in the R2A culture medium (R2A LAB163) (14).
3.3. Molecular Identification of Cyanobacterial Strains Based on 16S rRNA Sequence
3.3.1. DNA Extraction
The DNA extraction was done by the CTAB method (6). The qualitative method was used to determine DNA quality by loading on gel electrophoresis, and the quantitative method was done by NanoDrop Thermo spectrophotometer (Scientific).
3.4. Amplification of the 16S rRNA Gene Sequence
The PCR program included three cycles to amplify the 16S rRNA gene sequence. The first cycle was done at 94°C for 5 min. The second cycle included three steps of 94°C for 30 s, 56°C for 30 s, and 72°C for 2 min. The third cycle was done at 72°C for 15 min.
3.5. Screening for the Presence of NRPS and PKS Genes in the Identified Cyanobacterial Samples
The NRPS gene was amplified for adenylated domains (15). The PCR program for amplifying NRPS and PKS genes was designed in three cycles. Cycle one, the initiation step, was performed at 94°C for 5 min. Cycle two consisted of three steps: the first step consisted of unwinding the double-stranded DNA at 94°C for one minute, the second was ligation, in which the primers (Appendix 1 in Supplementary File) were attached to the single-strand template DNA at 51°C for one minute, and the third step was elongation at 72°C for 1.5 min. The third cycle included a step called final elongation, performed at 72°C for 7 min. Sequencing was performed to confirm that the amplified products were NRPS adenylated domains and PKS keto synthase domains.
A fragment of approximately 1 kb of adenylated NRPS and 700 bp from rRNA amplification was sequenced. Sequencing of PCR products was performed with an automatic Bechman sequencer by Microgene (South Korea).
3.6. Bioinformatics Analyses and Phylogenetic Tree Drawing
The resulting sequences were aligned by BioEdit software. Nucleotide blast and X-blast searches were performed to find similar sequences of the 16S rRNA gene and the NRPS and PKS genes available in the GenBank™ database at NCBI; hence, a consensus sequence was obtained.
One hundred strains that had the most similarity with the desired gene were aligned using the MAFFT VERGEN7 site, and finally, after selecting the best model, the phylogenetic tree was drawn using the Iq tree web server online program. Then, the tree was edited using the Figtree program. Gloeobacter violaceus strain VP3-01 (FR798924) was used as the root.
3.7. Prediction of Activated Amino Acid by Adenylated Module and Identification of Possible Conformation Type
Activated amino acid, signature sequence, and type of bioactive compound by NRPS A module were predicted by software downloaded from http://www.tigr.org/jravel/nrps (16, 17).
3.8. Drawing the Three-dimensional Structure of the Protein
To analyze the PKS protein sequence, the website (NaPDoS) https://npdomainseeker.sdsc.edu was used, and the predicted parameters were searched and suggested. In addition, the SBSPKSv2 website was used to analyze the phylogeny of PKS domains. This website modeled the structure of different PKS modules, got the PKS sequence, and performed BLAST. The proposed catalytic domains were modeled using homologous crystal structures from the PDB. The MODEL 3D PKS modeled the homology in the form of a biologically active dimer and the complete structure of the PKS module with individual catalytic domains. Hence, these models can be used as different catalytic domains to identify the vital residues involved in the domain interactions. The website https://swissmodel.expasy.org/ was used to analyze the protein sequences of the NRPS gene, and finally, the three-dimensional model of the structure was drawn. On this website, the accuracy, correctness, and reliability of the model protein named QMEAN, GMQE, and Ramachandran map were suggested.
4. Results
4.1. The Results of Morphological Identification
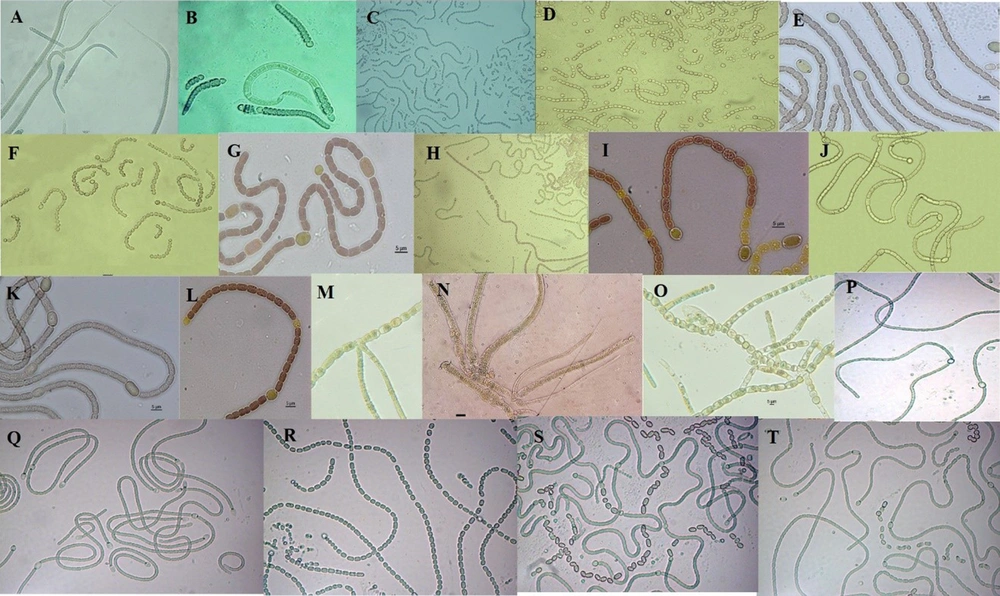
The results of morphological taxonomy investigations of cyanobacteria found in Tehran waterfall showed 20 strains (Figure 1) of different cyanobacteria belonging to three families Nostocacea, Scytonemataceae, and Rivulariaceae.
Native cyanobacterial strains found in Tehran waterfall; (A) Calothrix sp. F6; (B) Calothrix sp. alborz; (C) Nostoc sp. FA1; (D) Nostoc sp. FA3; (E) Nostoc sp. FA5; (F) Nostoc sp. F16; (G) Nostoc sp. F27; (H) Aliinostoc sp. F25; (I) Nostoc sp. F187; (J) Aliinostoc sp. bahar-M; (K) Nostoc sp. N66; (L) Aliinostoc sp. bahar-E; (M) Heteroscytonema sp. Ft11; (N) Heteroscytonema sp. F12; (O) Neowestiellopsis sp. F29; (P) Nostoc sp. F42; (Q) Nostoc sp. F17; (R) Nostoc sp. F19; (S) Nostoc sp. F3; (T) Nostoc sp. Ft salt.
4.2. The Results of Molecular Identification of 16S rRNA, NRPS, and PKS Genes
The results of three replications of PKS genes showed that only 20 strains belonging to the genera Nostoc, Calothrix, Aliinostoc, and Heteroscytonema contained polyketide synthase and nonribosomal peptide synthetase genes. The amplified genes were registered in the gene bank, and each one was assigned an access code, as specified in Table 1.
| Strains | 16S rRNA (Accession No) | NRPS (Accession No) | PKS (Accession Number) |
|---|---|---|---|
| Calothrix sp. F6 | MG549308 | MG548365 | MG548375.1 |
| Calothrix sp. alborz | MH014811 | KY548064.1 | KY548063.1 |
| Nostoc sp. FA1 | MG385055 | MG548370 | MG432824.1 |
| Nostoc sp. FA3 | MG385056 | MG548369 | |
| Nostoc sp. FA5 | MG385057 | MG432826.1 | |
| Nostoc sp. F16 | MG549316 | MG548379 | |
| Nostoc sp. F27 | MG549318 | MG432823 | MG54838 |
| Aliinostoc sp. F25 | MG549317 | MG548372 | |
| Nostoc sp. F187 | MG549311 | MG548377 | |
| Aliinostoc sp. bahar-M | MF803969 | JF795279 | JF795281 |
| Nostoc sp. N66 | KY548059 | KY548060 | KY548061 |
| Aliinostoc sp. bahar-E | MF803970 | JF795278 | MG432825.1 |
| Heteroscytonema sp. Ft11 | MG549309 | MG548366 | KY548067 |
| Heteroscytonema sp. F12 | MG549312 | MG548368 | MG548378 |
| Neowestiellopsis sp. F29 | MG385060 | MG432820 | |
| Aliinostoc sp. F42 | MG385059 | ||
| Aliinostoc sp. F17 | MG549310 | ||
| Nostoc sp. F19 | MG549313 | ||
| Aliinostoc sp. F3 | MG549314 | ||
| Desikacharya sp. Ft salt | MG549320 |
4.3. Results of Phylogenetic Tree Analysis Based on 16SrRNA Gene Amplification
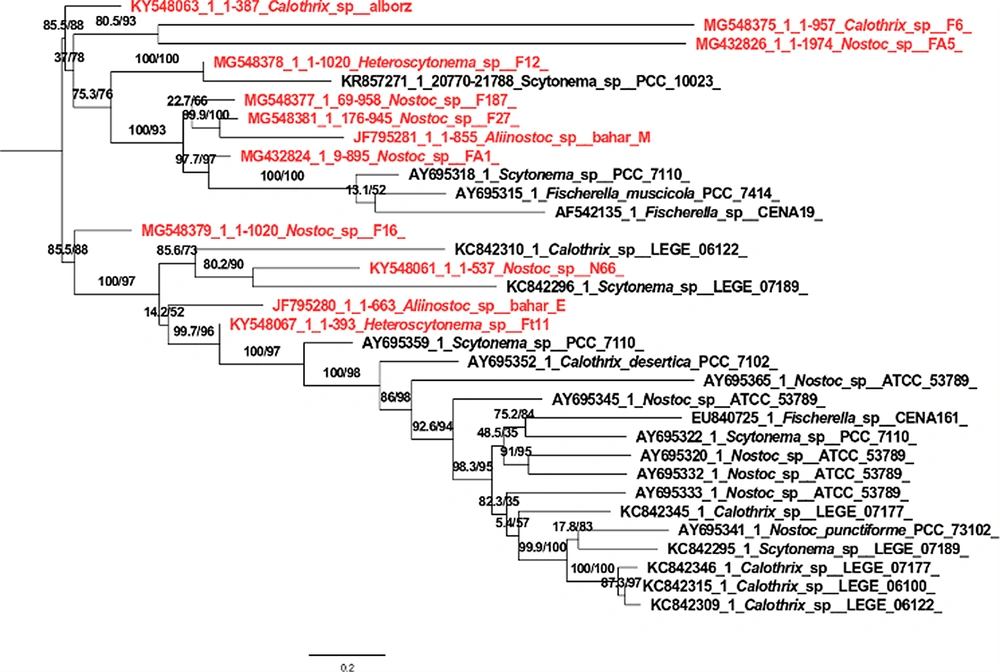
According to the phylogenetic tree made by the maximum likelihood method, each branch determines the relationships between taxa in terms of generation or ancestor. A node is formed from the connection of two branches, which is the place where lineages diverge. Nodes indicate the existence of taxonomic units (clade). The branch length indicates the number of differences compared to the common ancestor. The branch length indicates the number of changes in a branch. The studied strains are shown in red rank in Figure 2. The tree created by the Iq tree server method showed that the studied strain Calothrix sp. F6 had the most phylogenetic affinity with the Calothrix sp. 14SS strain, and they were placed in one clade with high Bootstrap support (91.77%). The Calothrix sp. Alborz strain had the most affinity with the Calothrix sp. PCC 7716 strain, and they were placed in one clade with high Bootstrap support (78.1%). The next clade included Heteroscytonema strains. The studied strain Heteroscytonema sp. Ft11 was placed in the same clade as the Scytonema sp. UCFS10 strain with high phylogenetic affinity and Bootstrap support (94.96%). The studied strain Heteroscytonema sp. F12 had the highest affinity with the strain Heteroscytonema sp. Ft11, and they were placed in the same clade with high Bootstrap support (90%). In the next clade, the studied strain Neowestiellopsis sp. F29 has the highest affinity with Bootstrap support (100%) which is included in a clade. The next clade included Nostoc strains, three strains containing Nostoc sp. FA3, Nostoc sp. FA1, and Nostoc sp. FA5. They were placed in a clade with high phylogenetic affinity and Bootstrap support (94.2%). In the next clade, the studied strain is placed in a separate branch with Nostoc eicroscopicum MBDU102 strain, with high Bootstrap support (69.5%). The last clade included Aliinostoc. Nostoc sp. F19 strain and Nostoc_muscorum_CCAP_1453_8, with 96.99% Bootstrap, Nostoc_sp__F27 strain and Nostoc_sp__SN64 strain with 98% Bootstrap, Nostoc_sp_F16 strain and Nostoc_sp__PCC_7120 strain with 82% Bootstrap, Nostoc_sp__F187, and Nostoc_sp__N66 strains with 100% Bootstrap placed in the same clade. Aliinostoc sp. F3 strain and Aliinostoc sp. Bahar_E strain had a phylogenetic affinity and placed in the same clade with Bootstrap support of 83.5%. Aliinostoc sp. Bahar_M strain and Aliinostoc_sp__F42 strain with Bootstrap support of 73.4%, Aliinostoc_sp__F17 strain, and Nostoc parmelioides SULS-P1 strain with Bootstrap of 96.6%, and Nostoc sp. PCC 8112 strain and Aliinostoc sp. F25 strain with a Bootstrap of 88.9% were placed in the same position.
4.4. Results of Phylogenetic Tree Analysis Based on NRPS Gene
The phylogenetic tree drawn by the maximum likelihood method of the strains containing the NRPS gene and other genes in the gene bank showed that the study strains were not genetically placed in the same clade based on phylogenetic relationships. In addition, the type of amino acid activated by the adenylation domain was not the same in the strains that were placed in the same clade. However, the predicted type of bioactive compound was reported to be the same in all studied strains placed in one clade with high bootstrap support.
In general, the results of phylogenetic relationships obtained from NRPS gene sequencing did not match the results of clade according to the 16S rRNA gene. Amino acid substrates, including serine, glycine, phenylalanine, threonine, aspartic acid, leucine, and tryptophan, are known as amino acid precursors for binding to adenylated domains of nonribosomal peptide synthetases. Serine amino acid was found in most adenylated amines of 12 cyanobacterial strains containing the NRPS gene (Appendix 2 in Supplementary File) (Table 2).
| Strains | Number of Domains | Signature Sequences | Predicted Amino Acid | Coordinate A3-A6 | E Value | Bioactive Compound |
|---|---|---|---|---|---|---|
| Aliinostoc sp. bahar_M | A-domain 1 | DVWFISLID- | NosA-M2-Ser | 2179 | 0.100 | Nostopeptolide synthetase |
| Aliinostoc sp. bahar_E | A-domain 1 | DVWHISLMD- | NosA-M2-Ser | 71236 | 0.017 | Nostopeptolide synthetase |
| Aliinostoc sp. F25 | A-domain 1 | DVWHISLID- | NosA-M2-Ser | 164 339 | 0.017 | Nostopeptolide synthetase |
| Nostoc sp. FA3 | A-domain 1 | DAWTVAGIC- | TycB-M3-D-/L-Phe/Trp | 129 330 | 0.037 | Nostopeptolide synthetase |
| Nostoc sp. F19 | A-domain 1 | DLFNNALTF- | McyA-M2-Gly | 66 268 | 1.6 | Nostopeptolide synthetase |
| Heteroscytonema sp. F12 | A-domain 1 | DLTKIGHVGK | SrfAB-M2-Asp | 117 338 | 0.034 | Nostopeptolide synthetase |
| Heteroscytonema sp. Ft11 | A-domain 1 | DVWHISLIDK | NosA-M2-Ser | 475 684 | 0.017 | Nostopeptolide synthetase |
| Calothrix sp. F6 | A-domain 1 | DLFNNALTY- | McyA-M2-Gly | 277 479 | 0.026 | Nostopeptolide synthetase |
| Desikacharya sp. Ft salt | A-domain 1 | DVWHVSL | PvdD-M3-Ser | 171 320 | 0.13 | Nostopeptolide synthetase |
| Heteroscytonema sp. F12 | A-domain 1 | DVWHVSL | PvdD-M3-Ser | 171320 | 0.13 | Nostopeptolide synthetase |
| Neowestiellopsis sp. F29 | A-domain 1 | DVWHISLID- | NosA-M2-Ser | 64273 | 0.017 | Nostopeptolide synthetase |
| Calothrix sp. alborz | A-domain 1 | DFWNIGMVH- | CchH-M2-Thr | 1184 | 0.006 | Nostopeptolide synthetase |
| Nostoc sp. N66 | A-domain 1 | DAWFLGNVV- | McyB-M1-Leu | 73278 | 0.008 | Nostopeptolide synthetase |
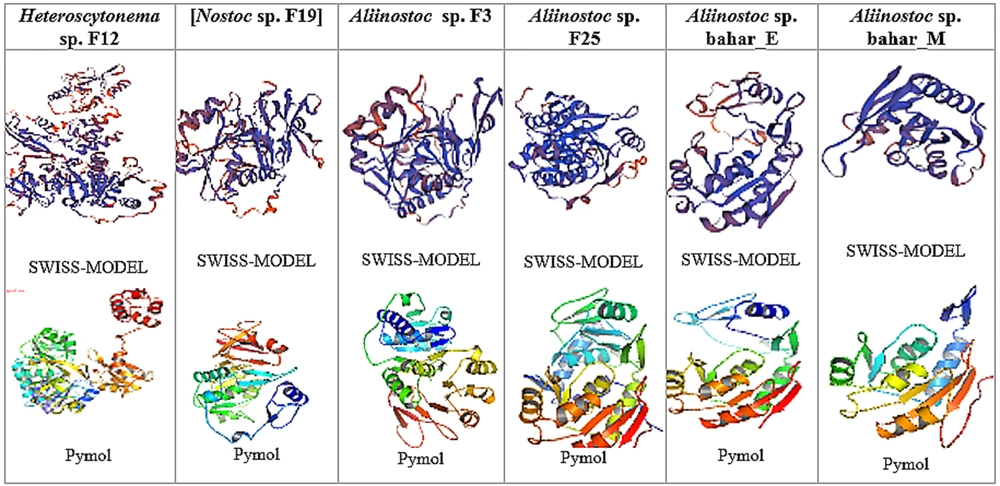
4.5. Results of Drawing the Secondary Structure of NRPS Gene Protein With SWISS-MODEL
The three-dimensional structure of the sequences obtained from the NRPS gene, belonging to the conserved adenylated domain of 12 investigated strains, was drawn using the conserved domains search site. The used web server selected the best model for 12 strains and presented the best structure using PDB and PyMOL software (Figure 3). The proposed model in the Swiss Model was evaluated based on QMEAN and GMQE. These indicators show the modeled protein's reliability, accuracy, and correctness in terms of the physical, chemical, and folding properties. As known, QMEAN is designed to evaluate the quality of the overall structure and the amino acid of the modeled structure. The QMEAN index includes four structural descriptions of rotation angle potential, beta carbon interaction, atom distance potential, and solvent coverage potential (18). Also, GMQE is a qualitative evaluation that combines the alignment features of the template model and shows a number between 0 and 1 (19). Evaluating the quality of the modeled structure after energy optimization and greater reliability of modeling through the Ramachandran map is a widely used method for protein rotation angles and evaluating the stereochemical quality of the modeled structure (20). It also shows that the protein can exist and have a natural structure biologically and naturally.
The size distribution of protein structure and the number of NRPS gene residues of 12 cyanobacterial strains are available in the diagram in Appendices 3 and 4 in Supplementary File. The size of the protein structure is shown as a star with overlapping points. In addition, the number of residues, along with the predicted percentage of similarity with the target protein in the form of open peaks with high deviation and the calculation of negative QMEAN, indicates the compatibility of the proposed model with the studied protein. The blue-purple diagram parts show the degree of similarity of the investigated protein with the proposed protein structure, and the red-orange parts show the degree of difference with the structure of the studied protein. Using different models to investigate the protein structure and Ramachandran's map confirms that over 90% of the amino acids are where they can be physically and chemically, showing the correctness of the predicted protein structure. Appendix 5 in Supplementary File shows the selected pattern, sequence identity, coverage rate, and the similarity percentage of the predicted pattern with the target peptide and the pattern selected in the SWISS-MODEL software.
4.6. Results of Molecular Phylogeny Analysis and PKS Protein Sequence
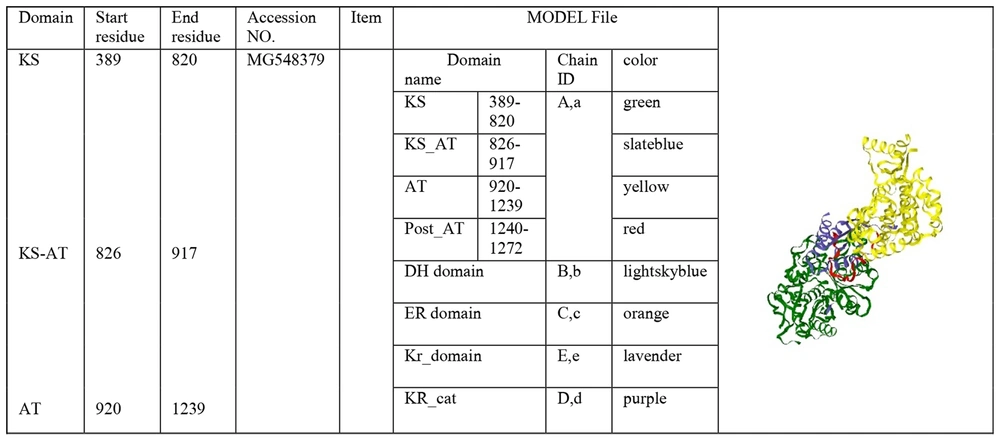
The phylogenetic tree of 12 strains containing the PKS gene is shown in Figure 4. The PKS protein sequence is located on this website http://nrps.igs.umaryland.edu/, where the different catalytic domains of the protein are shown. Clicking on each catalytic domain leads to the three-dimensional structure model, and clicking on the Jmol display part leads to a generalized three-dimensional structure model (Figure 5).
5. Discussion
Due to the abundance of natural ecosystems in Iran, conducting scientific research in various fields in these areas is necessary. For this reason, in this research, samples were taken from the running water of Tehran waterfall, and BG110 medium was used to increase the number and growth of cyanobacteria. In this research, 20 cyanobacterial strains were isolated, purified, and screened to search for gene-producing antibiotic compounds. Bioinformatics analysis and phylogenetic tree drawing were used to interpret the sequences.
Many researchers searched for polyketide synthase genes and nonribosomal peptide synthetases in cyanobacteria. Sequence analysis was done using bioinformatics software to investigate different modules that synthesize nonribosomal compounds and the role of each in amino acid activation (21, 22).
The strains with the NRPS and PKS genes were placed in a cluster with the strains of the same name in the phylogenetic tree with high support, showing their phylogenetic proximity. Clustering NRPS domains with taxonomic specificity between adenylated domains shows cyanobacterial genera and strains. For example, few samples had close relationships between taxonomic status and predicted composition, so the type of specific compound produced cannot indicate the genus type.
A phylogenetic tree drawn with the adenylated domain sequence of 12 strains together with other amino acid sequences showed differences between the genus type and the type of coding combination. For example, the combination of Nostopeptolde synthetase and the amino acid serine produced is not specific to a specific genus and may also be found in other cyanobacterial genera (Table 2). Among the genera found by BLASTx, the presence of amino acid substrates, including serine, glycine, phenylalanine, threonine, aspartic acid, leucine, and tryptophan as amino acid precursors, was shown for binding to the adenylated domain of nonribosomal peptide synthetases (Figure 4). The studied strains were not clustered in the phylogenetic tree with high support with the strains of the same name, which does not indicate their phylogenetic proximity. The clustering of PKS domains did not show taxonomic relationships between PKS domains among cyanobacterial genera and strains. This means that non-synonymous species were placed in a cluster or next to each other. Twelve studied strains had the same conserved domain. They are placed in a clade based on the characteristics of the protected domain. Ehrenreich et al. (15) showed no clear relationship between the biological activities and the presence of these genes and their biochemical and antibiotic activities in producing secondary metabolites (15). So, half of the tested strains had the PKS genes, while they had no specific antimicrobial activity.
Other conflicting results showed that the two strains of Lyngbya sp. PCC719 and Cyanothece sp., despite the presence of biological activity, lacked genes encoding natural products. These results suggest that the observed biological activities may be controlled by systems other than NRPS and PKS genes (21). The final confirmation of the relationship between the presence of NRPS and PKS genes and biological activities requires gene expression tests so that the relationship between biochemical activities and these unique genes, as well as the signaling system effective in the expression of these genes, will be known (23). Sequencing NRPS and PKS fragments encoding for adenylation and keto-synthase domains may lead to the prediction of NRPS substrate specificity and subsequent secondary metabolites (24-27), along with elucidating cryptic gene clusters (28, 29). Our analysis showed that clustering the partial A-domain is possible when substrate-conferring tendencies are taken into account, although in some cases, the amino acids are not homologous within the strains of each cluster.
These findings suggest that amino acid sequence identity to individual A and KS domains from characterized genes is not yet an applicable functional estimator for newly detected genes by NRPS and PKS PCRs within Cyanobacteria because of the scarcity of characterized Cyanobacterial NRPSs and PKSs. Besides, predicting functions based on the primary molecular structure will be critical for future discovery and rational engineering of polyketide synthase and nonribosomal peptide synthetase. Our results confirm those previously described by Ehrenreich et al. (15).
We could identify the specific branches for some amino acids. Analysis of A-domains' specificity towards particular amino acids shows motifs that recognize three amino acids. These amino acids are associated with forming five-membered heterocyclic rings, which form thiazole, oxazole, or their reduced structures in bacterial secondary metabolites. These signature pharmacophore compounds are common to many clinically important natural products, such as bacitracin, leinamycin (antibacterials), bleomycin, and epothilone (anticancers). The incorporation of these amino acids is done through a cyclization (Cy) domain that catalyzes amide bond formation, a cyclization of the side chain nucleophile with the newly formed amides, and a final dehydration (26).
5.1. Conclusions
Creativity in using different methods can lead to the evolution of new methods to get the maximum benefit from bioactive compounds. Although many investigations have searched the inhibitory effects of antimicrobial compounds, no information is available about the distribution of PKS and NRPS genes in Tehran waterfall in the literature. We conducted one of the first studies in this field in Iran. The recent isolation of new protease inhibitors and metabolites with antibiotic, antiviral, anticancer, and antifungal enzyme activities has made it more critical to use cyanobacteria as beneficial organisms in the pharmaceutical industry.