1. Context
Lung cancer (LC) is one of the most prevalent cancers worldwide after breast and colon cancers. LC accounts for 18% of all cancer-related deaths (1, 2). According to the World Health Organization (WHO), LC starts with genetic damage to DNA and epigenetic changes, which cause impaired normal cell function, cell proliferation, apoptosis, and DNA repair (3-6). The most prevalent symptoms of LC are hemoptysis, weight loss, dyspnea, and thoracic pain (7, 8). Lung cancer causes the death of many people worldwide every year. According to statistics from the WHO, 1.59 million people lost their lives due to LC in 2012. Also, about 1.79 million LC-related deaths will occur worldwide in 2020. Risk factors for LC include cigarette smoke, air pollution, radon, occupational exposures, and genetic factors. Tumor necrosis factor (TNF-α) is a genetic factor involved in LC (1, 9-12). Timely diagnosis is useful in reducing the death rate caused by LC. Researchers seek biomarkers for the timely prediction and diagnosis of LC (13). Tumor necrosis factor is a pro-inflammatory cytokine secreted by immune cells and plays an important role in the induction of apoptosis. Any disruption in the production of TNF-α is related to the cancer development of cancer (13-15).
A study showed a significant relationship between TNF-a-238 polymorphisms and the reduced risk of cervical cancer in Argentinian women (16). Another study revealed that TNF-a-238 polymorphisms were associated with an increased risk of liver cancer in the Asian population (17). Despite the significant findings regarding the positive association of TNF-a-238 polymorphisms with various types of cancer (13-15, 17), the results of some studies were not promising in this field (15, 18). Moreover, our knowledge indicates that no reports and definitive evidence exist about the association of TNF-a-238 polymorphisms with LC in human or animal models. Therefore, this study aimed to investigate the relationship between TNF-a-238 polymorphisms and LC was investigated in this research. The results of this study can be useful in the timely identification and design of treatment methods to improve LC in human samples.
2. Materials and Methods
2.1. Search Strategy
Articles were searched through Google Scholar, Scopus, and PubMed electronic databases until 2022 based on the keywords "Lung cancer," "238 Gene”, and “tumor necrosis factor.” The articles were selected based on the PRISMA flow diagram. Articles were searched based on keywords ("LC" OR non-Small CELL LC OR "CELL LC" OR "Small CELL LC") AND ("TNF-a-238" OR Tumor necrosis factor-238) AND (361625).
2.2. Selection Criteria for Articles
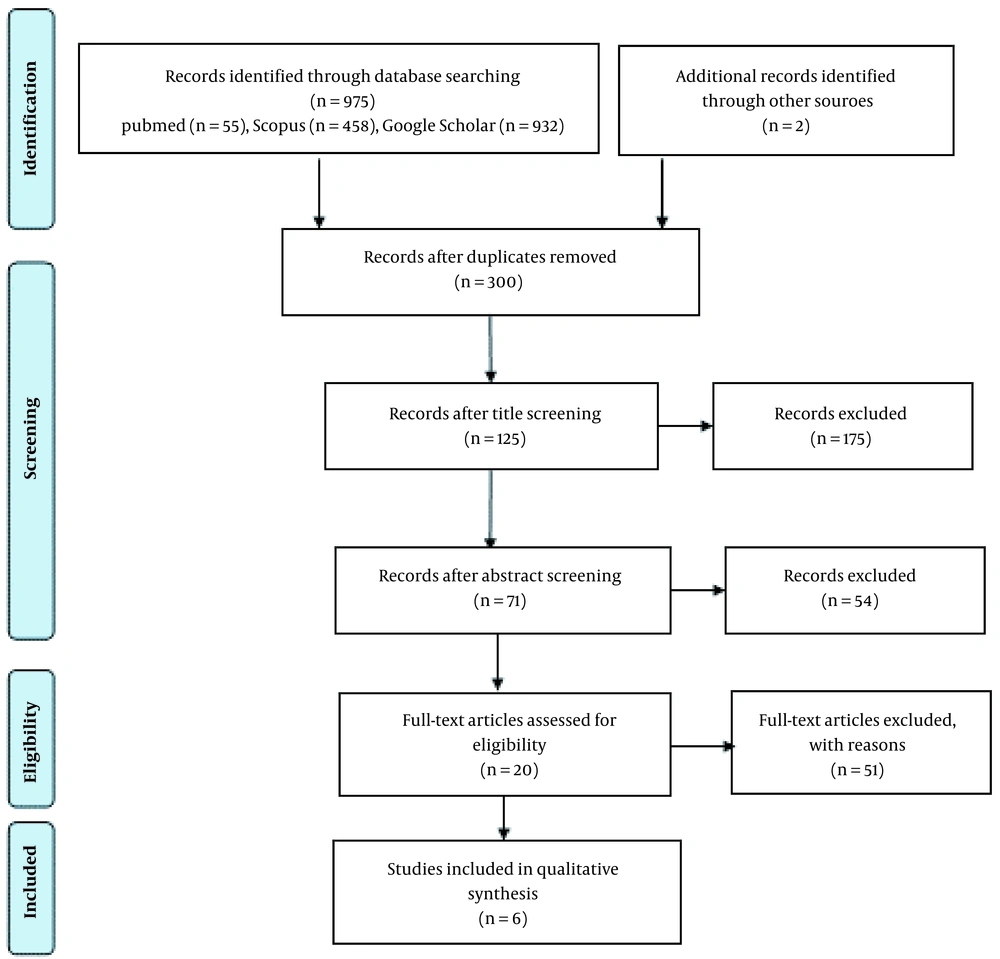
In the first stage, 975 articles until 2022 were entered into EndNote, of which then 300 articles remained by excluding duplicate articles. Next, 125 articles remained, and 175 were excluded from the study after screening the titles of the articles. By screening the abstracts of the articles and excluding 54 irrelevant articles, 6 articles that met the inclusion criteria were finally subjected to meta-analysis (Figure 1). Articles were selected based on the PRISMA flow diagram.
The studies were conducted on male and female adults in populations from Croatia, Taiwan, China, Tunisia, and the Americas between 2006 and 2018 (Figure 1).
Inclusion criteria for articles were human studies with the intervention of the effect of TNF-a-238 polymorphisms on LC lung cancer, English articles, articles that defined cancer patients with symptoms of hemoptysis, weight loss, dyspnea, and thoracic pain, and those with LC patients and a control (healthy) group. Finally, animal and in vitro studies, meta-analyses, reviews, and clinical studies were excluded from the study (19).
2.3. Statistical Analysis
Regarding the heterogeneity of the studies, they were merged using a random-effects model due to the heterogeneity of the studies, which was evaluated by Cochran's test and the I2 index. Data were analyzed using STATA software.
3. Results and Discussion
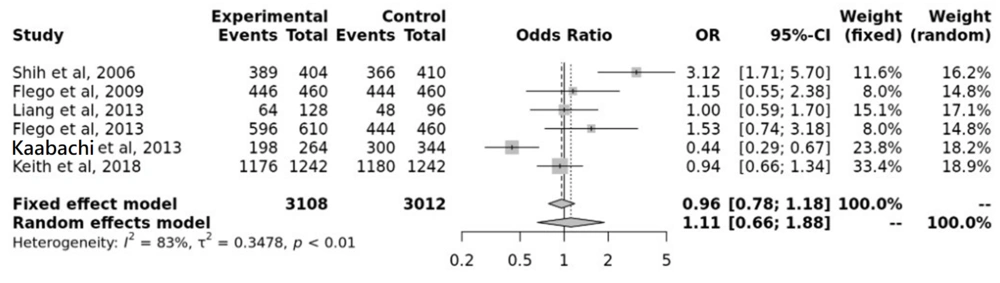
Based on electronic searches of several databases in this study, all publications related to the association between LC and rs361525 polymorphisms were investigated until 2022. The association of TNF-α-238 with the LC risk of LC was investigated in a wide range of populations using a meta-analysis. Table 1 lists the general characteristics of the meta-analysis articles and shows the distribution of the rs361525 genotype. The 95% odds ratio (OR) and 95% confidence interval (9% CI) for each study were determined to estimate and evaluate the effect and relationship between the TNF-α-238 gene and the risk of LCLC risk. The data on polymorphisms identified in each study showed the presence of heterogeneity. Therefore, the combined studies were merged using an additive model (AM). The results showed a significant difference between the two studies, while no significant difference was found in the other studies. However, the final overall OR reports with a value of (0.66; 1.88) OR = 1.11 indicate that the additive model AM in the TNF-α-238(rs361525) SNP increases the LC risk of LC in the random model (Figure 2).
| Authors | Year of Publication | Population | Method | Cases (n) | Controls (n) | Overall Sample Size | Count (Genotype) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-cancer Control | Lung Cancer | |||||||||||
| AA | GA | GG | AA | GA | GG | |||||||
| Shih et al. (14) | 2006 | Taiwan | PCR | 202 | 205 | 407 | 0 | 44 | 161 | 0 | 15 | 187 |
| Flego et al. (20) | 2009 | Croatia | PCR | 230 | 230 | 460 | 0 | 16 | 214 | 0 | 14 | 216 |
| Liang et al. (13) | 2013 | China | PCR-RFLP | 138 | 138 | 276 | 23 | 2 | 23 | 25 | 14 | 25 |
| Flego et al. (21) | 2013 | Croatia | PCR-RFLP | 27 | 174 | 201 | 0 | 0 | 98 | 0 | 0 | 6 |
| Kaabachi et al. (15) | 2013 | Tunisian | PCR | 133 | 174 | 307 | 2 | 40 | 130 | 14 | 38 | 80 |
| Eaton et al. (18) | 2018 | USA | PCR | 625 | 625 | 1250 | 2 | 58 | 561 | 1 | 64 | 556 |
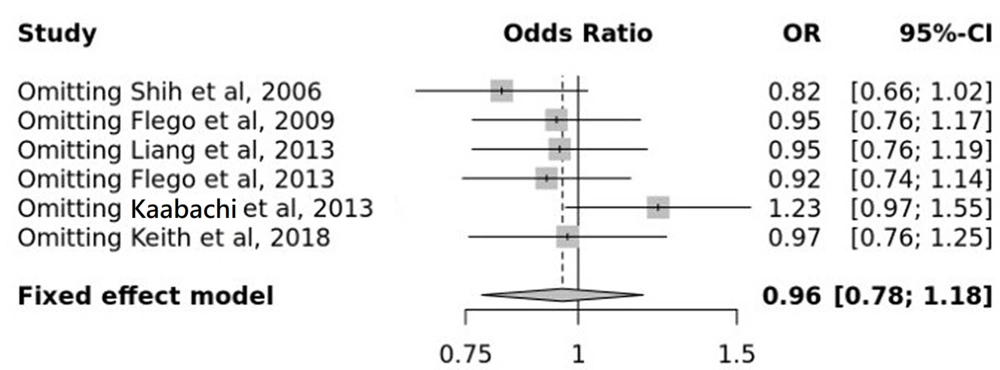
On the other hand, besides the entered data, the OR and the data analysis results show that the overall OR equals 0.96 (0.78; 1.18) with a 95% CI. Therefore, the rs361525 SNP reduces the chance of LC in the fixed model (Figure 3).
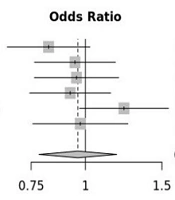
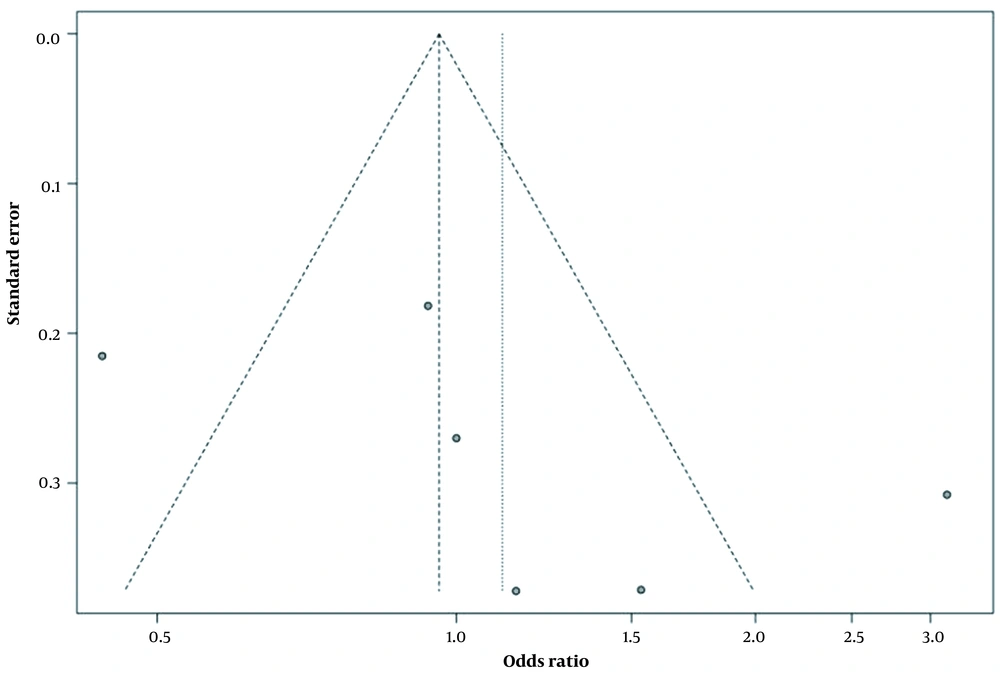
Figure 4 shows the results of distribution bias data shown symmetrically in the funnel diagram. As can be seen, there is (Figure 4) show no bias (P = 0.83) in the analyses, and there is also evidence of high heterogeneity between the studies (I2 = 83%, P = 0.83).
Although the relationship between the TNF-α-238 gene polymorphisms and various types of cancers is reported in many studies (13-17), the association between this gene polymorphism and LC is still one of the most challenging issues. Therefore, the present study investigated the association between TNF-α-238 polymorphisms and LC patients using database search methods. The results showed that the two studies differed significantly based on the obtained results, while no significant difference was found in the other studies. Moreover, the AM in the TNF-α-238 (rs361525) SNP increases LCLC risk. In agreement with this finding (Table 1), Liang et al. investigated the relationship between the genotypes of TNF-α-238 polymorphisms and LC by the PCR-RFLP method. They reported that the GG genotype of TNF-α-238 polymorphisms and the G allele could have an additive effect on LC in a Chinese population (13). Kaabachi et al. investigated the relationship between the genotypes of TNF-α-238G polymorphisms and LC using the PCR technique. They found that TNFα-238G > A could increase LCLC risk in the Chinese population (15). Also, Flego et al. examined the relationship between the genotypes of TNF-α-238 polymorphisms and LC using the PCR-RFLP technique. This study showed that the GG genotype of TNF-α-238 polymorphisms and the G allele could have an additive effect on LC (21). Flego et al. studied the relationship between the genotypes of TNF-α-238 polymorphisms and LC using the PCR-RFLP method. Their results indicated that the TNF-α-238 polymorphisms were not significantly related to the severity of LC (20). Shih et al. reported a significant relationship between the 238 G/A polymorphisms in the TNF promoter region and increased LC by the PCR technique (14). In contrast, the results of Eaton et al. presented evidence that TNFα-238G > A could reduce the risk of LCLC risk (18). A study showed a significant association between the TNF-a-238 polymorphism and the reduction of cervical cancer risk in Argentinian women (16). A relationship between TNF-a-238 polymorphisms and the increased risk of liver cancer in the Asian population was reported in another study (17). Despite the significant findings regarding the positive association of TNF-a-238 polymorphisms with various types of cancer (13-15, 17), the results of some studies are not promising in this field (15, 18).
In this study, since our search was limited to studies published in English, which may lead to some bias. Moreover, there were inadequate studies for the subgroup analysis. Therefore, more studies are needed to identify the precise association of TNF-a-238 with the increased risk of LCLC risk.
4. Conclusions
The results of this meta-analysis study show the relationship between TNF-a-238 and LC. TNF-a-238 polymorphisms increase the risk allele. TNF-a-238 with an OR = 1.11 has an additive effect on LC development and increases the risk of LCLC risk.