1. Background
Border disease (BD) is an important viral disease of sheep and goats. Barren ewes, abortion, stillbirth, and the birth of small, weak lambs that may suffer from tremors and abnormal body conformation are the most common demonstrations of BD. Infected animals have hairy fleeces. There are seven clusters for the border disease virus (1, 2). Border disease virus (BDV) transmission has been described among cattle, sheep, and goats (3, 4). The disease causes major economic losses in livestock due to reproductive failure and expensive inspections in livestock (5). The BDV genome is a linear ssRNA, and the viral proteins include 5'-Npro, C, Erns, E1, E2, p7, NS2-3, NS4A, NS4B, NS5A, and NS5B-3' (6). As known, NS3 possesses highly conserved domains and plays essential roles during the virus life cycle (7).
An important difference between CP and NCP BDV is the expression of NS3 in CP-BVDV (8). The NS2–3 cleavage is vital for pestiviral replication since NS3 possesses serine protease, RNA helicase, and nucleoside triphosphatase (NTPase) and replicate activities that cannot be done by uncleaved NS2-3 (9). The N-terminal one-third of NS3 has a protease activity and cleaves the BDV polyprotein. The C-terminal domain of NS3 is necessary for BDV replication (10).
As conventional therapies for treating human and livestock viral diseases have limitations and alternative treatments are urgently needed, gene therapies such as RNAi technology have been applied as potential therapeutic measures. As a plus-strand RNA virus, BDV appears to be an attractive target for siRNA or shRNA since its genome functions as both a replication template and mRNA (11). A common method to evaluate the effectiveness of gene therapy is to monitor cell lines expressing target genes of gene therapy. For this purpose, the first step is to clone the desired target sequences in a suitable vector to prepare a cell line highly expressing that gene fragment. Due to the conservation of BDV-NS3 and its importance for viral protein synthesis, it has been frequently chosen as a target. Thus, in the current work, the conserved helicase and nucleotide binding site domain of BDV-NS3 was selected for cloning in a lentiviral plasmid (11-13).
Lentiviral vectors (LVs) have been widely used as gene therapy vectors in antiviral therapies in recent years. The LVs based on HIV-1 have become an ideal vector for gene transfer works. They can integrate and transfer genes into dividing and non-dividing cells such as neurons, lymphocytes, and macrophages. The replacement of the env gene with VSV envelope G protein expands the range of the host cells (14). Three or four plasmids enter the cell separately in lentiviral packaging systems, so the chance of producing reproducible lentiviruses after recombination is low (14, 15). We selected pCDH-CMV-MCS-EF1-cGFP-T2A-Puro as a cloning vector because of its availability and having cutting sites of routine restriction enzymes and resistance genes to available antibiotics. Furthermore, this vector has a green fluorescent protein gene that helps evaluate transfection efficiency. Strong RSV, CMV, and EF1 promoter sequences are included in this plasmid.
2. Objectives
The present research aimed to construct a recombinant lentiviral plasmid expressing HELICc and nucleotide binding site domain of BDV-NS3 as a transfer vector.
3. Methods
3.1. Preparation of Helicase Fragment of BDV-NS3
The X818 BDV- NS3 helicase domain sequence was obtained from National Center for Biotechnology Information (NCBI) database and then aligned with all known gene sequences in other BDV strains extracted from NCBI. The alignment was performed using the online software www.ebi.ac.uk/Tools/msa/clusalo. Then, EcoRI and BamHI cutting sites were included in the 5' and 3' terminals, respectively. Furthermore, the sequence of His Tag was added to the 3' end. Shine Gene Molecular Biotechnology Company (China) synthesized the designed fragment and cloned it in pUC57.
3.2. Restriction Enzyme Digestion
For the insertion of the desired fragment, pCDH lentiviral plasmid (Addgene) and pUC57 carried BDV- NS3 helicase was digested using BamHI (Roche, Germany, Cat. No. 10220612001) and EcoRI (Roche, Germany, Cat. No. 10220566001). The digestion reaction included 2 μg/μL of vector, 10 U of each enzyme, 10x K buffer, and deionized sterile water. Incubation was performed at 37°C for one hour, and one microliter of the product was electrophoresed to confirm the process.
3.3. Extraction of BDV-NS3 Helicase Fragment from Agarose Gel
To purify the digested plasmid, after digestion with EcoRI and BamHI and removing the undigested plasmids and restriction enzymes, Expin Gel SV (50 prep) kit (Gene all, Cat. No. 000001422) was applied to extract the cut plasmid and to insert DNA fragment from the agarose gel.
3.4. Ligation of BDV-NS3 Helicase Fragment Into Lentiviral Plasmid and Transformation
For the ligation of the insert fragment into the vector, 2 μL of T4 10x ligation buffer (Fermentas, Germany, Cat. No. B69), 2 μL of 50% PEG-4000 (Fermentas, Germany), 1 μL of T4 DNA ligase (Fermentas, Germany, Cat. No. EL0013), 150 ng of inserting fragment, 50 ng of plasmid mixed with nuclease-free water up to 20 μL total volume were prepared. Heat shock protocol was applied for the transformation of the ligation mixture. In summary, 10 μL of ligation product was added to 100 μL of DH5α (F-endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG purB20 φ80dlacZΔM15 Δ(lacZYA-argF) U169, hsdR17(rK-mK+), λ-). Then, the microtube was placed in the following conditions: 30 min at 4°C, 90 s at 42°C, and 5 min at 4°C. Afterward, 900 μL of LB broth was inserted into the mixture and shaken at 200 rpm for 45 min. Afterward, the mixture was centrifuged at 6000 rpm for 3 min. Finally, the pellet culture in LB agar +100 mg mL-1 ampicillin was performed and put overnight at 37°C.
3.5. Colony PCR
The PCR and Mini preparation method of plasmid extraction was performed for ten individual clones of each plate. The PCR was done with the general primers of vector (CMV-F: AATGGGCGGTAGGCGTGTA -3'and EF1-R: 5'- GGACTGTGGGCGATGTG -3'). The PCR thermal cycling conditions consisted of 94°C, 5 min for one cycle, 30 cycles at 94°C for 30 s, 55°C for 40 s, 72°C for 60 s, and the final step at 72°C for 8 min. The positive BDV- NS3 helicase fragment was synthesized and cloned in the pUC57 vector by Shine Gene Molecular Biotechnology Company (China), and negative controls (deionized water) were also tested. The PCR included 6.5 µL of DEPC water, 5.5 µL of Taq DNA Polymerase 2x Mix Red-MgCl2 1.5 mM (Amplicon, Cat. no. A180301), 1 pmol of reverse and forward primers, and 50 ng of extracted DNA up to 15 µL total volume.
3.6. Sequencing
After electrophoresis in 0.8% agarose gel, two reactions of colony PCR-positive samples containing the recombinant plasmids were sent to Bioneer Company (Korea).
4. Results
4.1. Extraction of BDV-NS3 Helicase Fragment from Agarose Gel
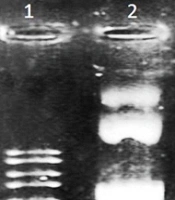
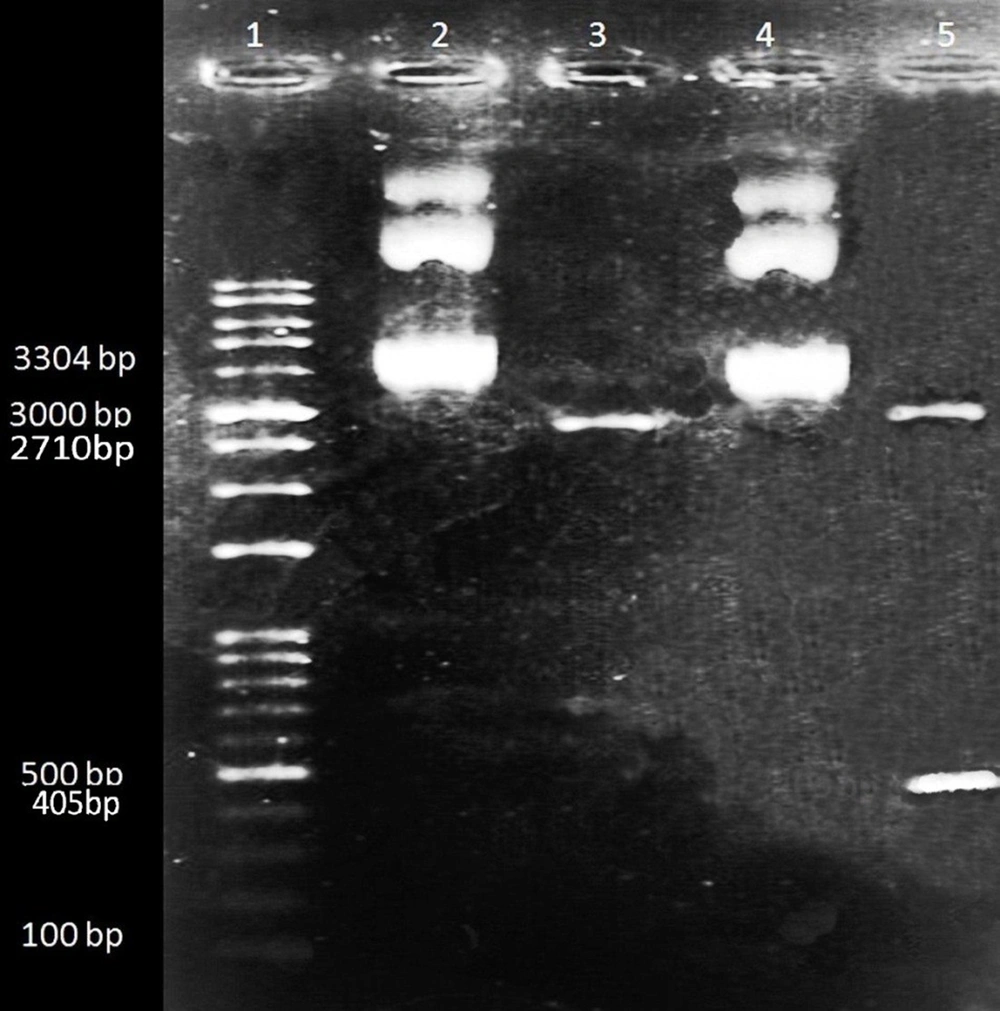
One microliter of cut vector and one microliter of the undigested plasmid were run in 0.8% gel electrophoresis near an Excel Band™ 1 KB Plus (0.1 - 10 kb) DNA marker and the extraction of the fragment from digested pUC57, and the difference of size between cut and uncut plasmids was observed (Figure 1).
Extraction of the digested and uncut pUC57 plasmids. 1: 1 kb DNA Ladder; 2: Uncut pUC57 carrying BDV-NS3 helicase fragment (3304 bp); 3: Digested pUC57 plasmid (2710 bp); 4: Uncut pUC57 carrying BDV-NS3 helicase fragment (3304 bp); 5: Upper band: Cut pUC57 (2710 bp) and lower band: BDV-NS3 helicase fragment (405bp)
4.2. Colony PCR
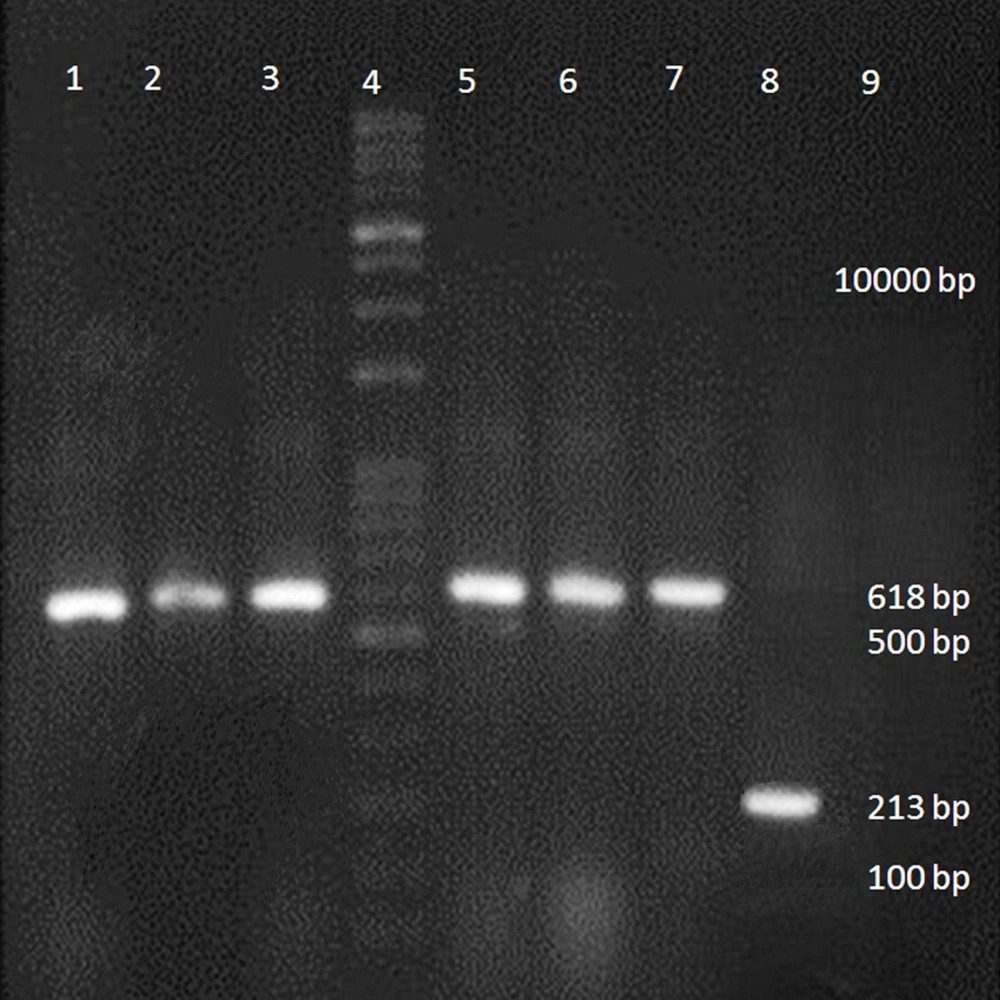
The BDV- NS3 helicase was visible in agarose gel with a size of 618 bp (Figure 2). The band related to the pCDH plasmid was 213 base pairs in size. To confirm that these primers did not bind to the genome of DH5α. The PCR was also performed for untransformed DH5α, and no band was observed (negative control).
4.3. Extraction of BDV-NS3-Helicase- pCDH-CMV-MCS-EF1-cGFP-T2A-Puro
One microliter of extracted plasmids from two positive clones was electrophoresed on 1% agarose gel, and the production of the recombinant vector was confirmed by observing the desired band at 8602 bp (Figure 3).
4.4. Sequencing
Analysis of the read sequences of recombinant plasmids showed the desired fragment's presence and the cloning process's accuracy (Figure 4).
5. Discussion
Pestiviral diseases have a great impact on livestock production. Because border disease, common in sheep populations, is not genetically similar to common BVDV strains, the BVDV vaccine is unsuitable for cattle for use in sheep (4, 16). Unlike BVDV vaccines, the use of anti-BDV vaccines is low, and the vaccines produced are inactivated. Attenuated live vaccines or recombinant subunits for BDV are not commercially available. Pestivirus contaminants of modified live virus vaccines may cause a serious disease following their inoculation in domestic ruminants (17).
Genetics has been involved in treating pestiviruses in recent years due to the low efficiency and adverse off-target effects of chemical compounds. New studies have been developed on the antiviral gene therapy approaches against these viruses. Accordingly, designing and developing methods for evaluating such treatments is necessary. Therefore, the present study performed the early stages of developing an approach to assess genetic treatments against BDV. One way to evaluate the effectiveness of such molecules is to evaluate them in cell lines expressing the genes they want to suppress. To achieve this goal, the first step is to clone the desired sequences to evaluate the expression change in a suitable vector for transfer to a suitable host cell (18).
In studies performed for gene therapy against members of the pestiviruses, the NS3 gene has been selected as the target because of the special role of the NS3 protein in the cleavage of the polyprotein and the advancement of the pathogenicity goals of the virus. Therefore, this gene was also selected for cloning into a lentiviral plasmid in the present study. The HELICc and nucleotide binding site conserved domain was chosen. Mokhtari et al. produced MDBK cells expressed BVDV-5′UTR and BVDV-NS3 using lentiviral vectors (19), used single and multiple siRNA molecules opposed to NS3 CSFV, and showed that viral isolates from different subtypes are inhibited by this method (20). In the present study, for permanent expression of the target genes, the transduction was performed by a lentiviral vector. Plasmids have also been used for gene expression induction after transfection, although the duration of gene expression is low. Lentiviral vectors induce remarkably sufficient gene expression in their host cells for a long time. Furthermore, LVs possess effective delivery because they potently integrate into the host chromatin, eliminate all the pathogenic genes in the vector, and finally have no interference with the pre-existing antiviral reactions of the immune system. Furthermore, LVs can transmit large nucleotide sequences (3000 bp), and the probability of mutagenesis and carcinogenesis is low after their application (21-23).
Given the reasons mentioned above, lentiviral vectors were selected to induce the target gene expression in appropriate cells, and at first, the desired nucleotide fragment was cloned in the lentiviral transfer vector.
As known, pCDH-CMV-MCS-EF1-cGFP-T2A-Puro, selected in the present study, has ampicillin and puromycin resistance genes, both of which are easily and abundantly accessible. On the other hand, this plasmid has a green fluorescent protein gene, which helps us as a marker to evaluate the accuracy of transfection. In addition, strong RSV, CMV, and EF1 promoters are included in the plasmid to express the target gene and GFP. The multiple cloning sites in this plasmid include the cleavage sites of the most restriction enzymes (24, 25), firstly increasing the potency of LVs (pCDH-CMV-MCS-EF1-cGFP-T2A-puro) and epEGFP-N1 for expression of HIV-1Nef protein (26, 27). Ranjibar et al. amplified the Cop-GFP from the pCDH-CMV-MCS-EF1-cGFP-T2A-Puro plasmid for developing a phagemid for the production of mammalian reporter cells (28).
In the last two decades, many studies have been performed using interfering RNA molecules to reduce gene expression to identify gene function or antiviral therapy against viruses, including Flaviviridae. However, there is no such study for BDV. Some research evaluating anti-BVDV RNAi has been described since BVDV has a high structural and biological similarity to BDV. More recently, BVDV-1 proliferation has been inhibited by siRNAs that target 5'-UTR, (C), NS4B, and NS5A, and a specific evaluation method has been employed by producing a cell line expressing BVDV subgenomic replicons (12). Mishra et al. showed the suppressive effects of siRNAs targeting envelope genes and the 5ʹ-UTR of BVDV. In this study, gene expression suppression was monitored using subgenomic replicon-expressing cells (11). Ni et al. described an shRNA system that effectively inhibits BVDV replication. They prepared BVDV subgenomic replicon expressing cell lines to evaluate shRNAs (13).
Unfortunately, there is no study about preparing lentivectors carrying BDV genes. Several cell lines expressing genes or subgenomic replicons of some other Flaviviridae members have been generated and are used to evaluate treatments based on the repression of gene expression. For example, Puig-Basagoiti et al. prepared a cell line expressing the Western Neil's virus sub-replicons after cloning the target gene segments into appropriate plasmids and transfecting them. These cells were successfully used to assess the chemical inhibitors of the WNV epidemic strains (29). Kumar et al. developed a lentivirus-based shRNA system targeting envelope genes to suppress JEV and WNV (30). Furthermore, Kumar et al. showed that pseudotyping the lentivirus with RVG enabled the delivery of the transferred gene to the neurons in a cell-specific manner with a lower dose of lentivirus (31). Henry et al. used lentiviral viruses that expressed NS3, NS5b, and IRES from 5'UTR of HCV to infect the Huh-7 cells (22).
The results of the present study showed that the BDV-NS3 target fragment was successfully cloned in the desired plasmid, and it will be accessible for transfection of the appropriate cell lines using lentiviral packing systems to produce specific reporter cells.