1. Background
Phosphorylation and dephosphorylation of proteins are vital post-translational modifications, acting as a reversible change to control different functions of proteins, such as protein-protein interaction and protein localization in response to different stresses such as drought, salt, and cold (1). It is indispensable to probe into the identification and functional description of the PP2C gene family, cementing the base for understanding its essential molecular mechanism in stress signaling (2, 3). Protein phosphorylation is one of the important steps that helps regulate some physiological and biochemical reactions. PPs carry out dephosphorylation. Plant protein phosphatase 2Cs (PP2Cs) are important in plant hormone signaling, growth processes, and environmental stress responses (4). The high proportion of PP2C genes reveals their evolutionary consequence, prerequisite, and participation in various plant and cellular functions. For instance, it is demonstrated that TaPP2C genes are associated with wheat's developmental stages and stress responses (5). The high proportion of PP2C genes indicates their evolutionary significance, requirement, and involvement in diverse plant cellular functions (6). The PP2C genes have been termed a regulator of water deficit tolerance and act as a negative regulator in the ABA-signaling in plants (7). Transgenic studies confirmed the expression level of the ZmPP2C10 gene as a negative regulator in drought stress tolerance in maize (5). Evaluating two mutants, abi1-1 and abi2-1, indicated that two PP2C genes are involved in different physiological processes after exposure to abiotic stimuli containing salt, drought, and cold (8).
Gene duplication, critical for gene family development, is considered a type of genome expansion formed through block and tandem duplication (9). Paralogous gene pairs located on the same chromosome are considered as tandem duplication. Gene duplicates are prevalent in plants and, in some cases, contribute to evolutionary novelty. Gene duplication allows the detection of paralogs, where their presence in the genome leads to identifying the gene family.
2. Objectives
This study identified 61 HvPP2C genes from the barley genome and grouped them into 12 subfamilies. Comprehensive analyses of functional annotation, gene duplications, chromosomal distribution, and phylogeny of these HvPP2Cs were further carried out. To validate in silico analysis, their expression profiles were also investigated by qRT-PCR under cold and heat treatments in 3 different tissues (root, stem, leaf). The results presented here provide a foundation for further functional characterization of HvPP2C genes in this model species.
3. Methods
3.1. Physicochemical Characteristics, Functional Annotation, Phylogenetic Analysis, and Chromosomal Distribution of the HvPP2C Genes
As explained earlier, two techniques were utilized to detect putative HvPP2C genes in barley. In the first technique, a protein homology search with accessible PP2C proteins from Arabidopsis and rice was performed. The second technique included retrieving the PP2C protein sequence using hidden Markov model (HMM) analysis, with Pfam number PF00481 containing the tubule normal domain from the Pfam HMM library. The Arabidopsis and rice protein sequences were taken from TAIR and RAP-DB databases. The known Arabidopsis PP2C protein sequences were taken from NCBI and utilized as query sequences for the tBLASTn program in barley to search for similar protein sequences. All putative sequences were approved with the SMART database and interproscan. The remaining 10 non-redundant candidates were recognized as HvPP2C proteins. The ExPASy protoparam (www.expasy.org) was used to calculate the theoretical isoelectric point (pI) and molecular weight (Mw) of each HvPP2C protein. The protein sequences of HvPP2Cs were uploaded to the online annotation tool of Mapman (www.plabipd.de/portal/mercator-sequence-annotation) for functional annotation and categorization using default parameters. HvPP2C protein sequences were aligned using the ClustalW function of MEGA 7.0. Then, the phylogenetic tree between barley, rice, field mustard, and Arabidopsis was determined using the neighbor joining (NJ) algorithm with 1000 repeated bootstrap test parameters. To locate HvPP2C genes on barley chromosomes, HvPP2C genes were placed on each chromosome. HvPP2C genes were mapped on all chromosomes with MapChart (10).
3.2. Detection of Paralogous and Prediction of TFBS HvPP2C Genes in Barley
Similar gene pairs to HvPP2C proteins were identified by Blastp with more than 85% identity from Ensembl Plants. Similar genes to HvPP2C were visualized using the Circos program (http://mkweb.bcgsc.ca/tableviewer/visualize/). After determining the genomic sequence of each gene, 2000 bp upstream of the transcription start region of HvPP2C was recovered. Transcription factor binding site (TFBS) prediction was also done using the PlantCARE website.
3.3. Barley Growth Under Heat and Cold Treatments
To study the gene expression of HvPP2Cs genes under abiotic stress, seeds of Azaran and Jolge cultivars were grown in peat moss-filled pots and were kept under greenhouse conditions: Day/night temperatures were maintained at 25°C, with a 16/8 h light/dark period. Young root stems and leaves from the 2-week-old seedlings were harvested for tissue-specific expression analysis under cold and heat stresses and treated for four hours at cold (4°C) and heat stress (42°C). Different plant tissues were harvested 4 hours after abiotic stresses and immediately stored at −80°C for further analysis. Barley cultivars that grew normally were used as normal replicates. All experiments were repeated 3 times.
3.4. RNA Extraction and Quantitative Real-time PCR Analysis
Total RNA was extracted from roots, stems, and leaves of cold and heat under stress conditions at the seedling stage four hours after heat and cold treatments using the RNA-Plus kit (Sinaclone, Iran). The purity and concentration of RNA were determined by NanoDrops, and its quality was confirmed using 1% agarose gel analysis. Then, cDNA synthesis was performed according to the instructions of the Easy cDNA Synthesis Kit, Iran. Three replicates were performed to analyze each gene. Barley actin gene was used as a reference gene. Gene expression of five HvPP2C were analyzed under cold and heat stresses. All primers used in gene expression analysis are listed in Table 1. Primers were designed based on HvPP2C genes using the Oligo program. Real-time PCR (qPCR) was performed on an ABI 7500 using SYBR Green Supermix as described in the manufacturer's instructions. Relative expression was determined through the delta-delta 2-∆∆Ct technique. RT-qPCR was performed to determine the expression profile of five HvPP2C genes using different tissues under heat and cold stresses.
| Primer Name | Sequence (5' - 3') | Annealing Temperature |
|---|---|---|
| Hvpp2c16 | 58 | |
| F | CAGGGACGGGGATCAAGTTGT | |
| R | GCCAGAGTCACTTGCACGA | |
| Hvpp2c37 | 60 | |
| F | TGCCGTGCTCCTACCATGA | |
| R | CATCCCTCTCTGTCCTCTGTC | |
| Hvpp2c70 | 59 | |
| F | TGTGGCAAATGCCGGGGATTC | |
| R | CCACCAGCGTTCTGAATCCTCTC | |
| Hvpp2c41 | 59 | |
| F | CACACTGCTGAGCTAGTTGTCC | |
| R | CTTCACGAGTTACCTGCAAC | |
| HvPP2C42 | 60 | |
| F | TGTGCGTAGAACCACCGCA | |
| R | CGACTGCATGGCTCATAGGATTC | |
| HvActin | 58 | |
| F | GGTCCATCCTAGCCTCACTC | |
| R | GATAACAGCAGTGGAGCGCT |
Primers and Their Sequences Were Used for This Study's Gene Expression Analysis of HvPP2C
4. Results and Discussion
Based on the bioinformatics analysis, the 61 HvPP2C genes were detected. These proteins' detailed information (biochemical properties) are listed in Table 2. Sequence analysis showed that the deduced HvPP2C proteins' length varied from 51 amino acids (HvPP2C6a) to 1266 amino acids (HvPPC61). The predicted MW and pI ranged from 4.56 kDa (HvPP2C36) to 141.01 kDa (HvPP2C61) and from 4.6 (HvPP2C6) to 9.26 (HvPP2C56), respectively (Table 2). When the domain and motifs of HVPP2C proteins are similar, they will be located in the same subfamily, suggesting functional similarities for the genes in the same subfamily (9).
| Uniprot accession | Protein Name | Number of Amino Acids | Molecular Weight (MW: kDa) | Theoretical pI | Genome Location | Duplication |
|---|---|---|---|---|---|---|
| M0USW5 | HvPP2C46 | 582 | 63.33 | 6.36 | chr1H:11138897-11142264 | Tandem duplicate |
| F2EHQ5 | HvPP2C47 | 298 | 32.76 | 4.71 | chr1H:54286009-54288770 | No duplication |
| A0A287FKC7 | HvPPC13 | 399 | 43.09 | 6.67 | chr1H:386658800-386662971 | No duplication |
| A0A287FL77 | HvPP2C72 | 392 | 43.32 | 8.23 | chr1H:390690911-390698376 | No duplication |
| A0A287FT97 | HvPP2C48 | 248 | 27.25 | 9.51 | chr1H:431534223-431538146 | Tandem duplicate |
| F2DJX7 | HvPP2C51 | 395 | 41.58 | 6.32 | chr1H:545790189-545793081 | No duplication |
| A0A287GS68 | HvPP2C52 | 534 | 57.95 | 4.89 | chr1H:552521506-552527955 | No duplication |
| A0A287IKQ2 | HvPP2C41 | 399 | 43.19 | 8.68 | chr2H:573578815-573582822 | Tandem duplicate |
| AK357653 | HvPP2C41a | 305 | 33.17 | 6.34 | chr2H:573626499-573629830 | Tandem duplicate |
| AK357622 | HvPP2C42 | 386 | 41.88 | 7.1 | chr2H:622771403-622774870 | Block duplicate |
| A0A287JAG6 | HvPP2C68 | 390 | 42.8 | 6.9 | chr2H:704204949-704207601 | No duplication |
| M0Y7N6 | HPP2C45 | 268 | 29.30 | 4.93 | chr2H:722484027-722488008 | Tandem duplicate |
| M0Y7N6 | HPP2C45a | 284 | 30.91 | 4.93 | chr2H:722563005-722566934 | Tandem duplicate |
| A0A287KAQ4 | HvPP2C01 | 353 | 37.42 | 5.68 | chr3H:46958934-46961758 | No duplication |
| A0A287KXD0 | HvPP2C03 | 333 | 35.67 | 7.99 | chr3H:291032153-291037688 | No duplication |
| A0A287KZD6 | HvPP2C57 | 331 | 35.72 | 6.67 | chr3H:340435807-340459284 | No duplication |
| A0A287L0C7 | HvPP2C6 | 400 | 41.41 | 4.6 | chr3H:360092700-360098013 | No duplication |
| A0A287L3R5 | HvPP2C7 | 390 | 45.93 | 6.7 | chr3H:407373335-407383431 | No duplication |
| A0A287LGB2 | HvPP2C6a | 51 | 5.51 | 9.81 | chr3H:511604930-511605643 | No duplication |
| A0A287LGC6 | HvPP2C18 | 241 | 25.79 | 5.35 | chr3H:511676340-511679818 | No duplication |
| M0ZC70 | HvPP2C16 | 393 | 41.86 | 5.78 | chr3H:614881000-614883250 | Block duplicate |
| A0A287MS52 | HvPP2C17 | 311 | 34.37 | 6.42 | chr3H:690155525-690157207 | No duplication |
| A0A287MUL9 | HvPPC17a | 385 | 42.74 | 8.97 | chr3H:699024910-699050008 | No duplication |
| A0A287NGV0 | HvPP2C36 | 317 | 4.56 | 7.59 | chr4H:98736380-98739103 | No duplication |
| A0A287NWL4 | HvPP2C75 | 394 | 43.47 | 6.07 | chr4H:358054764-358061365 | No duplication |
| A0A287P371 | HvPP2C21a | 402 | 44.04 | 5.26 | chr4H:449217161-449223939 | No duplication |
| M0UT12 | HvPP2C33 | 362 | 38.59 | 5.61 | chr4H:456797020-456800448 | No duplication |
| A0A287P934 | HvPP2C30 | 400 | 43.06 | 5.68 | chr4H:507379979-507384625 | No duplication |
| M0YIW1 | HvPP2C28 | 399 | 43.78 | 9.3 | chr4H:616241933-616249757 | No duplication |
| M0VSN9 | HvPP2C31 | 613 | 67.35 | 6.25 | chr4H:645763892-645766806 | No duplication |
| A0A287QIS3 | HvPP2C78 | 383 | 41.99 | 9.06 | chr5H:79975025-79980048 | No duplication |
| A0A287RMK2 | HvPP2C69 | 429 | 46.86 | 5.78 | chr5H:519091103-519096484 | No duplication |
| A0A287S6R9 | HvPP2C70 | 396 | 42.70 | 4.96 | chr5H:582294665-582301638 | Block duplicate |
| A0A287SF58 | HvPP2C34 | 265 | 28.55 | 8.85 | chr5H:603081538-603088903 | No duplication |
| A0A287SKH9 | HvPP2C21 | 273 | 30.22 | 5.43 | chr5H:620396504-620402041 | No duplication |
| A0A287SPR2 | HvPP2C58 | 423 | 46.87 | 6.1 | chr5H:632185424-632196887 | No duplication |
| BAJ98729.1 | HvPP2C61 | 1266 | 141.015 | 7.54 | chr5H:640812970-640820023 | Tandem duplicate |
| A0A287SS11 | HvPPC35 | 518 | 54.96 | 5.3 | chr5H:641283216-641287419 | No duplication |
| A0A287SV91 | HvPP2C36a | 286 | 31.55 | 5.36 | chr5H:648725477-648728034 | No duplication |
| A0A287TI85 | HvPP2C55 | 323 | 33.94 | 4.72 | chr6H:54801305-54811859 | No duplication |
| A0A287TKK2 | HvPP2C10 | 360 | 38.32 | 4.98 | chr6H:67703547-67706602 | No duplication |
| F2E5E2 | HvPP2C11 | 380 | 41.89 | 5.58 | chr6H:117906254-117913932 | Block duplicate |
| M0ZD44 | HvPP2C12 | 354 | 37.86 | 5.5 | chr6H:210762786-210767184 | Tandem duplicate |
| A0A287U0S8 | HvPP2C65 | 537 | 59.84 | 5.35 | chr6H:219263779-219288759 | Tandem duplicate |
| A0A287U288 | HvPP2C14 | 515 | 55.400 | 6.53 | chr6H:246534205-246538678 | No duplication |
| A0A287U425 | HvPP2C15 | 449 | 47.87 | 6.07 | chr6H:290392640-290399019 | No duplication |
| A0A287U5N1 | HvPP2C13 | 391 | 41.87 | 5.25 | chr6H:307794396-307808554 | No duplication |
| A0A287UBR2 | HvPP2C23 | 317 | 34.33 | 5.48 | chr6H:393388996-393389949 | No duplication |
| F2EHY7 | HvPP2C26 | 597 | 65.12 | 5.34 | chr6H:463066303-463070536 | No duplication |
| A0A287VGJ4 | HvPP2C37 | 372 | 38.86 | 4.85 | chr7H:11079479-11102169 | Tandem duplicate |
| A0A287VW12 | HvPP2C6b | 293 | 32.33 | 6.72 | chr7H:54028669-54033782 | Tandem duplicate |
| A0A287VVL1 | HvPP2C54 | 353 | 38.47 | 5.48 | chr7H:55007477-55011109 | Tandem duplicate |
| M0VNX5 | HvPP2C66 | 521 | 56.67 | 5.21 | chr7H:172453523-172458271 | No duplication |
| A0A287X173 | HvPP2C56 | 367 | 39.46 | 9.26 | chr7H:467679707-467683931 | No duplication |
| A0A287X5E1 | HvPP2C22 | 365 | 39.19 | 4.93 | chr7H:512082503-512087461 | No duplication |
| A0A287X703 | HvPP2C24 | 319 | 33.92 | 5.44 | chr7H:524549530-524550486 | Tandem duplicate |
| A0A287X703 | HvPP2C24a | 320 | 33.93 | 5.45 | chr7H:524639090-524640046 | Tandem duplicate |
| A0A287X703 | HvPP2C24b | 330 | 33.92 | 5.65 | chr7H:524814869-524815825 | Tandem duplicate |
| A0A287XCZ5 | HvPP2C76 | 848 | 96.84 | 5.74 | chr7H:570046163-570059714 | No duplication |
| F2D7E1 | HvPP2C58 | 391 | 42.55 | 4.97 | chr7H:636403133-636408251 | No duplication |
| F2E1Q6 | HvPP2C60 | 392 | 43.53 | 8.52 | chr7H:646432525-646437065 | No duplication |
Characteristics of HvPP2C Proteins Found in the Barley Genome
4.1. Phylogenetic Analysis, Chromosomal Distribution, and Duplication of HvPP2C Genes
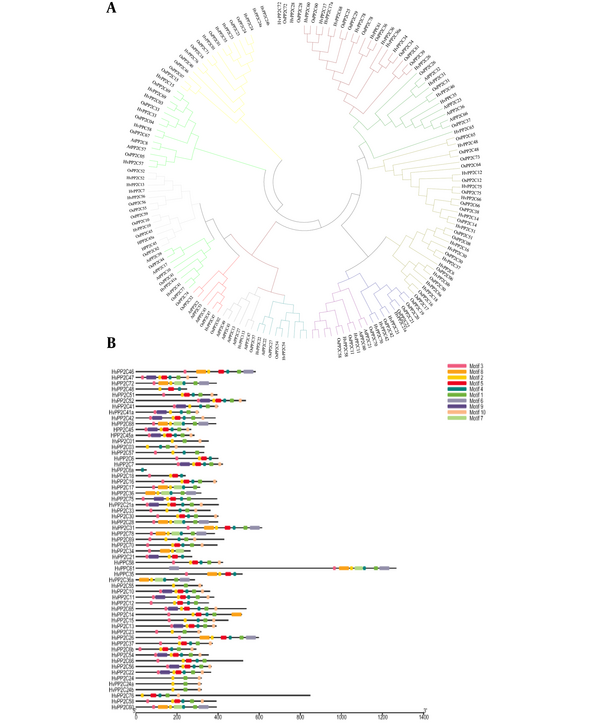
To assess the phylogenetic relationships of the 61 HvPP2C proteins in barley, we performed a phylogenetic analysis using MEGA7 software (Figure 1). The phylogenetic tree showed that the PP2C genes can be divided into 12 subgroups. Previous studies showed that MtPP2C genes in Arabidopsis and rice were divided into 13 subfamilies (9, 11, 12). Some researchers have revealed that HvPP2C genes from monocots can be grouped into 13 subfamilies based on their domains (9, 13). The results show that the exact function of the genes of the subfamilies is due to the conserved motifs.
Most HvPP2C proteins were classified in the same subfamily (Figure 1B). Most proteins from different species were placed in the same cluster, had similar conserved motifs and functional activity, and were paralogous (14). Motifs 1, 2, and 3 were available in most subfamilies, and motif 1 was present in 61 HvPP2C proteins except for HvPP2C58 and HvPP2C60 (Figure 1B). These outcomes propose that the precise functions of various subfamily genes may result from conserved motifs.
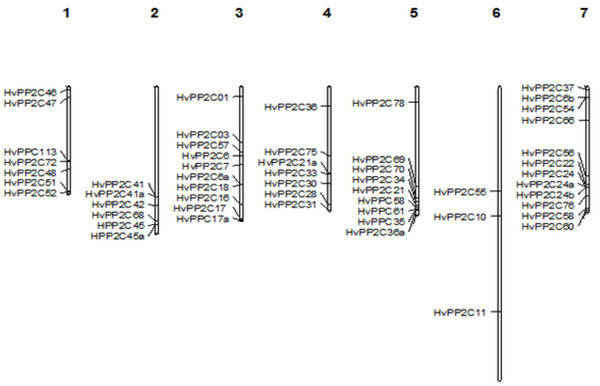
Afterward, the 61 HvPP2C genes were drawn using the MapChart software, and sixty-one HvPP2C genes were located across all seven chromosomes, ranging from 3 to 12 per chromosome (Figure 2). The number of HvPP2Cs on each chromosome varied, and chromosome 7 contained the largest number of HvPP2C members with 12 genes. However, the least number of genes was detected on chromosome 6, containing only three HvPP2C genes. In addition, seven HvPP2C were located on chromosomes 1 and 4. The 6 and 10 HvPP2C genes were located on chromosomes 2 and 3, respectively. Finally, nine HvPP2C were distributed on chromosome 5. Based on the research on rice, Arabidopsis, and B. distachyon, it has been shown that PP2C gene families mainly expanded through whole-genome and chromosomal segment duplications (9, 15). As shown in Fig 2, these 19 pairs of duplicated HvPP2C genes are distributed on chromosomes 1, 2, 3, 5, 6, and 7 but not on chromosome 4 (Figure 2).
4.2. Paralogous Genes Study and Gene Duplication in HvPP2C
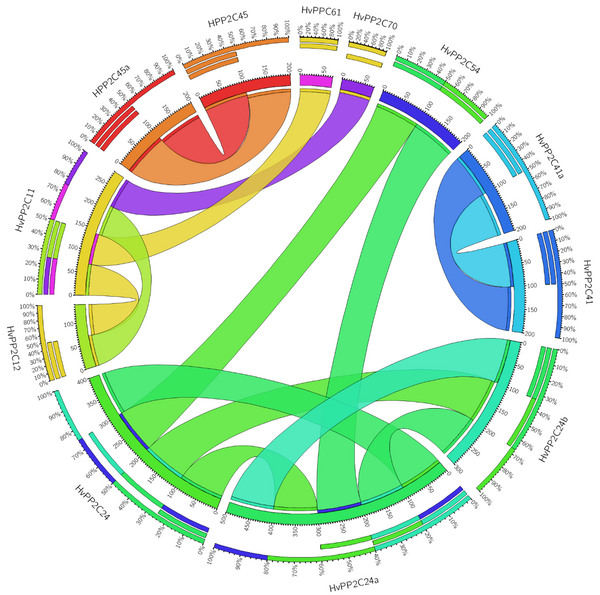
This study used synteny analysis to detect paralogous HvPP2C genes in the barley genome (Figure 3). Based on the results, five barley genes showed high similarity (identity 80%) and were implicated in paralogous. The analysis of synteny revealed that HvPP2C24b with HvPP2C24a, HvPP2C24 with HvPP2C24a and HvPP2C24b, HvPP2C5 with HvPP2C5a, and HvPP2C41 with HvPP2C41a were paralogous. It was observed that tandem duplicates occurred within HvPP2C24b, HvPP2C24, HvPP2C24a, HvPP2C37, HvPP2C6b, HvPP2C12, HvPP2C61, HvPP2C65, HvPP2C46, HvPP2C45, HvPP2C45a, HvPP2C48 and HvPP2C54 genes. Also, block duplication occurred within HvPP2C11, HvPP2C16, HvPP2C42, and HvPP2C60 genes. Analysis of HvPP2C indicated that genome duplication and tandem duplication play key roles in barley genome enlargement (16).
4.3. Prediction of TFBS in the HvPP2C Genes
In the promoter regions of HvPP2C genes in barley, TFBs, TCA, and ARE-responsive elements were investigated. In this study, TFBS included ARE (anaerobic responsive elements), TCA elements, TATA-box, and CAAT-box in promoter regions (Appendix 1). HvPP2C17a and HvPP2C37a genes had the highest TFB in their promoter regions (Appendix 1). Previous studies have shown that HvPP2C responds to various stresses (which stress) and stimuli in Arabidopsis, tomato, and soybean (9, 17). ARE regulatory elements are important for the induction of anaerobic respiration (11). Enrichment of the TCA element in the promoter regions of most HvPP2C genes suggests comprehensive transcriptional regulation by HvPP2C itself, indicating a complex regulatory network among them. According to a previous report, TCA elements (salicylic acid elements) have also been associated with abiotic stresses (18). This may be due to the presence of cis-elements in the promoter region of these genes (19). Our findings demonstrated that HvPP2C, HvPP2C37, and HvPP2C37 genes have multiple binding sites in the promoter regions and play key roles in response to various environmental stresses. Results also showed that HvPP2C genes regulate many important developmental processes in plants, such as seedling and reproductive stages, and are involved in abiotic resistance, such as heat and cold stresses.
4.4. Functional Annotation
In barley, the largest percentage of detected proteins was implicated in protein metabolism (80%), and the second largest class was involved in hormone metabolism (8.57 %) (Figure 4). The percentage of mitochondrial 2-oxoglutarate/malate carrier protein (misc) was 7.14%. Most of the HvPP2C genes in the barley were involved in protein metabolism (protein degradation, post-translational modification, and protein synthesis) and hormone metabolism. Different kinds of HvPP2Cs were involved in protein degradation-related proteins, implying that the PP2Cs might play major roles in protein degradation during the barley seedling stage. Our findings showed that HvPP2C genes play a key role in abiotic stresses like cold and heat stresses. The HvPP2C had TFBs responsive to hormonal stress, indicating its role in response to abiotic stresses.
4.5. Expression Profiles of HvPP2C Genes 3 Tissues Under Cold and Heat Stress Conditions
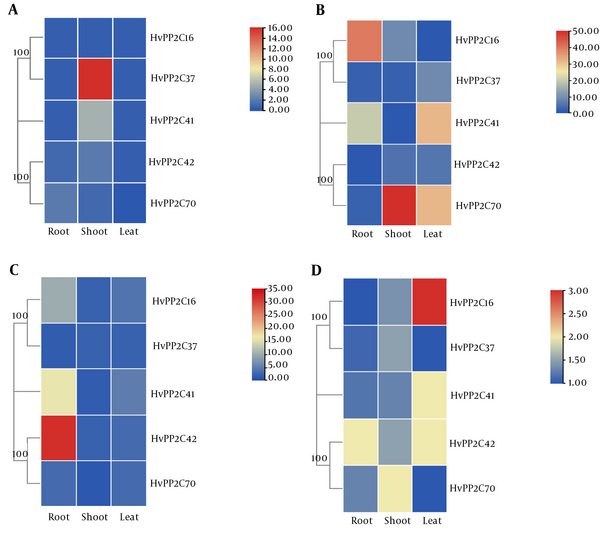
In Azaran and Jolge cultivars, the expression levels of five HvPP2C were determined under heat and cold stress conditions, most of which show a wide range of expression. To evaluate the expression profile of 5 PP2C genes and to evaluate the expression profile under heat and cold stress conditions, we utilized qRT-PCR analysis in different tissues: root, stem, and leaf. In the Azaran cultivar, HvPP2C37 and HvPP2C70 genes showed low expression in the root under cold stress conditions (Figure 5A), whereas HvPP2C16 and HvPP2C42 genes showed increased expression in the stem tissue (Figure 5A). In the leaf tissue, an increased expression of HvPP2C37, HvPP2C41, HvPP2C42, and HvPP2C70 genes was also shown (Figure 5A). Similarly, the expression levels of MtPP2C genes, such as MtPP2C37, were altered significantly during cold stress (9). In the Azaran cultivar, the HvPP2C gene showed decreased expression, except for the gene HvPP2C70, which showed increased expression in the stem tissue under cold stress (Figure 5A). Also, HvPP2C37 and HvPP2C41 genes were up-regulated in stem tissue under heat stress (Figure 5B). All HvPP2C genes showed reduced expression in leaves under heat stress (Figure 5B). Maximum expression was observed for the HvPP2C16 gene in the root compared with the other genes. It has been reported that among the BdPP2Cs studied, BdPP2C37 was increased under heat and cold stresses. Previous study has shown that BdPP2C70 strongly enhanced expression levels in response to abiotic stress, indicating that they may contribute in response to ABA (10). These results showed that the HvPP2C16 gene can be used as a molecular marker in barley improvement.
Differential gene expressions under cold (A) and heat (B) stress conditions in Azaran cultivar. The Jolge cultivar shows gene expression under cold (C) and heat (D) stress conditions. Blue and red indicate up and down-regulated genes, respectively. The yellow color indicates no significant expression under stress conditions.
The expression profiles of HvPP2C genes under heat and cold stresses revealed differential and overlapping expression patterns. In the Jolge cultivar, only the HvPP2C42 gene showed increased expression in response to cold stress in the root (Figure 5C), whereas the HvPP2C70 gene showed increased expression in response to heat stress in the shoot tissue (Figure 5D). Further, HvPP2C16, HvPP2C41, and HvPP2C42 genes were expressed in the leaves (Figure 5D). On the other hand, HvPP2C16, HvPP2C41, and HvPP2C42 genes were increased in the root in response to heat stress (Figure 5D). In the leaf tissue of the Jolge cultivar, HvPP2C16, HvPP2C41, HvPP2C42, and HvPP2C70 genes showed increased expression under heat stress (Figure 5D). Various expression patterns of HvPP2C genes may suggest their various roles in response to heat and cold stress conditions. In contrast to our results, the MtPP2C41 gene was decreased in response to the cold stress in Medicago truncatula (9).
5. Conclusions
In this study, 61 HvPP2C genes were detected in barley, and we surveyed their phylogenetic relationships, chromosomal locations, gene duplications, functional annotation, and prediction of TFBS. Also, bioinformatics analysis of these HvPP2C genes and their gene expression profiles were performed. Our results showed that the genes and groups with similar protein motifs had similar origins and possibly similar functions. These results provided insights into the evolutionary relationships of HvPP2C in the barley. HvPPT2C genes had different functions, indicating the presence of conserved domains in these genes. Based on the HvPP2C gene expression pattern, the HvPP2Cs gene showed different patterns in response to abiotic stresses. These results showed that the HvPP2C16 gene can be used as a molecular marker in barley improvement. However, the function of other subfamily PP2C in plant resistance to abiotic stress is poorly understood and needs further investigation. The results of our study establish a foundation for future studies on the functions of HvPP2C genes in plant’s response to abiotic stresses and provide a basic understanding that may allow us to elucidate the potential functions of HvPP2C genes under cold and heat stress conditions in barley.