1. Background
Coxiella burnetii, an intracellular bacterium, can cause query (Q) fever-related illnesses in humans and animals (1, 2). This Gram-negative bacterium can be easily distributed in different places, except for specific regions, such as New Zealand and Antarctica.
This bacterium is a primary source of infection in humans, domestic and wild animals, and birds. Despite its ubiquity, the epidemiology of this bacterium remains unclear, especially in poor source regions (3).
Query fever can be transmitted to humans mainly by farm animals such as cattle, sheep, and goats. In addition, different animals might be infected by C. burnetii, including horses, dogs, pigs, camels, ducks, geese, turkeys, water buffalo, pigeons, many wild birds, squirrels, deer, mice, harvest mice, cats, rabbits, and rats. The epidemiology of C. burnetii varies in different countries (4, 5). An infected animal can transmit C. burnetii through its urine and feces or respiratory system (6, 7). Contaminated raw milk typically causes more concerns since it can be considered an infection source in humans. Recently, several studies have been conducted globally in this regard, and their results revealed that C. burnetii may contaminate unpasteurized milk with an infection rate of 4.7 - 47.7% (8, 9). Hence, as recently reported, unpasteurized milk and other dairy products should be carefully examined to consider the C. burnetii infection before the products reach the consumer.
Indirect immunofluorescence, complement fixation, and enzyme-linked immunosorbent assay (ELISA) are the standard serological techniques for the detection of C. burnetii (10, 11). Furthermore, C. burnetii isolation is not normally expected in veterinary medicine and is not recommended as a systematic method since its implementation is relatively time-consuming and challenging. Moreover, this method requires a bounded level-three laboratory (12). Ruminants, including domestic water buffalo (Bubalus bubalis), are usually the primary and significant livestock species worldwide due to their high-quality milk, meat, and leather products. Due to their interaction, these animals' contact with wild or domestic animals, especially cattle, and their interactions with other ecosystems make them susceptible to various infectious diseases (13). Ruminants, including 800 000 cows and 160 000 buffalos, dairy, and meat products, are the primary income source for rural households in West Azerbaijan province, Iran (14, 15). Generally, milk can be regarded as the primary pathogen source or, more specifically, a pathogenic bacterium. This has been recognized as a substantial vector of pathogens since the prevalence of miscellaneous epidemics such as Staphylococcus aureus, C. burnetii, Mycobacterium bovis, and Salmonella spp. during the past decades (16). The C. burnetii can be easily excreted into nature by using infected livestock products such as milk. Hence, as mentioned earlier, nonpasteurized and contaminated milk can be the primary way to infect the consumer (16). C. burnetii may be transmitted by the ticks of certain species (17). Previous studies demonstrated that Hyalomma anatolicum anatolicum and kennel ticks are loaded with C. burnetii in Iran; however, the tick species was not examined (18).
Control, prevention, management, and treatment of Q fever in humans and animals require accurate and early detection of C. burnetii. Former studies on the occurrence of C. burnetii in dairy cows were mostly oriented by serologic tests to discover the antibodies introduced months earlier (6). It is very dangerous and complicated to isolate C. burnetii. Recently, C. burnetii was detected using polymerase chain reaction (PCR). It is a sensitive, safe, and specific procedure to detect C. burnetii in various specimens (19). Various target genes were used (20) for specific C. burnetii identification, such as com1 encoding a 27 kDa outer membrane protein, the superoxide dismutase (Sod B) gene, and the heat shock operon that encodes 2 heat shock proteins (htpA and htpB). The other target genes include the macrophage infectivity potentiator protein (cbmip), isocitrate dehydrogenase (icd), and a transposon-like repetitive region of the C. burnetii genome.
The com1-based PCR method has been proven to be a highly beneficial and sensitive method for detecting C. burnetii in different blood samples (21, 22). The com1 gene expresses and encodes a 27 kDa outer membrane-associated, immune-reactive protein and is remarkably preserved among C. burnetii isolates considered in different medical and terrestrial origins (22).
2. Objectives
The aim of the present study was to evaluate the presence of C. burnetii in milk samples collected from cattle and buffalo during warm and cold seasons in West Azerbaijan province, Iran.
3. Methods
The current investigation was conducted in West Azerbaijan province. This province is located in the northwest of Iran (37° 33′ 10.08″ N, 45° 4′ 33.24″ E). The wet winds of the Mediterranean and Atlantic oceans have a crucial impact on the weather of Urmia, the capital of West Azerbaijan (https://www.britannica.com/place/Azerbaijan-region-Iran) province. Furthermore, cold winds from the north can be the main reason for harsh winters. The related data are illustrated in Figure 1.
3.1. Milk Sampling
Overall, 600 milk samples were collected randomly from 74 dairy farms, with 300 samples from buffalo and 300 samples from cattle. Then, 10 mL of milk was collected from the udders of animals from different geographical regions of Urmia during four seasons in 2020 and placed into sterile vacuum tubes. The milk samples were categorized into 3 animal age groups (≤ 6, 7 - 10, and > 10 years). Samples from each group were kept on ice for immediate transfer to the Microbiology Laboratory at the Faculty of Veterinary Medicine.
3.2. Extraction of DNA from Milk Samples
Following the method presented by Parisi et al. (4), DNA extraction was performed on the milk samples that had already been subjected to centrifugation at 5000 rpm for 10 minutes. After discarding the fat from the samples, the samples were used in the DNA extraction process. DNA extraction was accomplished using the Blood Genomic DNA Extraction Mini Kit (50 preps, FAVORGEN, Taiwan) according to the manufacturer's instructions. The amount and quality of the extracted DNA were checked using the NanoDrop 2000c (Thermo Scientific, USA), and it was kept at -20°C until PCR was performed.
3.3. Molecular Identification of Coxiella burnetii by Using Nested PCR
Nested PCR targeting the com1 gene was required to detect C. burnetii molecularly. The applied primers in this study (Table 1) were similar to those used in previous studies conducted by Parisi et al. and Zhang et al. (4, 22).
| Gene Detected | Primer | Sequence 5-----3 | Amplicon Length (bp) | PCR Condition |
|---|---|---|---|---|
| Com-1gene | Omp1 | AGTAGAAGCATCCCAAGCATTG | 501 | 94 C for 120 s,10 cycles, 53 - 63 (touchdown), 94 C for 30 s, 53 C for 30 s, 72 C for 60 s, 25 cycles |
| Omp2 | TGCCTGCTAGCTGTAACGATTG | |||
| Omp3 | GAAGCGCAACAAGAAGAACAC | 438 | 94 C for 30 s, 53 C for 30 s, 72 C 60 s, 36 cycles | |
| Omp4 | TTGGAAGTTATCACGCAGTTG |
The first step of this method employed Taq DNA Polymerase Master Mix RED (Amplicon, Denmark). The PCR reaction was performed in a volume of 25 μL, consisting of 5 μL of extracted DNA, 50 picomoles of each primer (com 1 & com 2), and 12.5 μL of the Master Mix. Additionally, the touchdown (TD) PCR was used to optimize and improve the sensitivity of the reaction, which reduces pollution and inhibitors. The TD- and nested PCR thermal plans (Quanta Biotech, England) were performed based on the thermal cycler reported by (22).
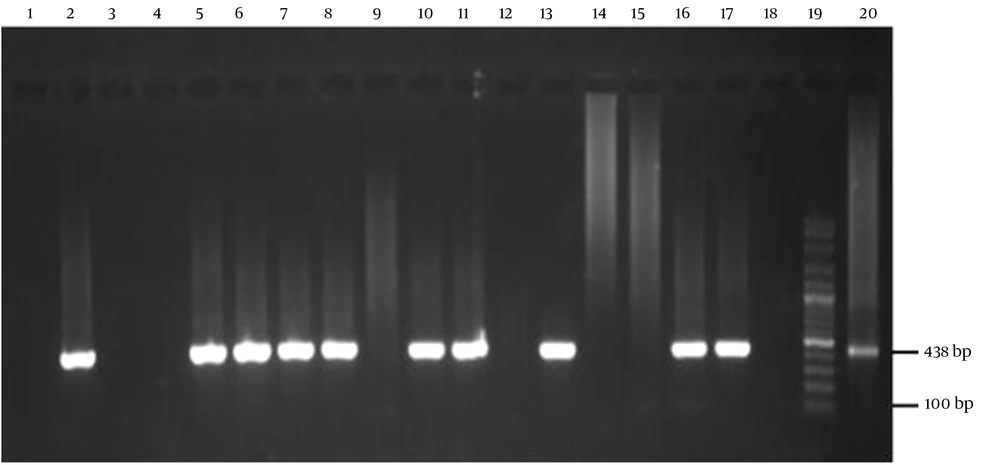
At this stage, following the previously explained nested PCR, the PCR was prepared, except for the DNA template. Furthermore, the temperature and thermal cycle conditions were based on Zhang et al.'s (22) study. Finally, the products obtained from each step of the PCR were electrophoresed on a 1.5% agarose gel containing a safe stain. Subsequently, they were visualized with InGenius Gel Documentation (Syngene Bio Imaging, United Kingdom) according to Figure 2.
Agarose gel image of the amplified fragment of C. burnetii com1 gene (438 bp) using nested polymerase chain reaction (PCR). Lane 3, 4, 9, 12, 14, 15 negative samples. Positive control is lane 2, 20 (nine mile strain), 100-bp molecular ladder in lane 19 (Smobio Technology Inc., Taiwan); 5, 6, 7, 8, 10, 11, 13, 16, 17 lanes positive samples, lanes 1, 18 negative control
3.4. Statistical Analysis
The chi-squared test was utilized to statistically analyze the data in SPSS v. 22 (IBM Corp., Armonk, NY, USA), and P < 0.05 was assumed as a significant criterion.
3.5. Nucleotide Diversity and Phylogenetic Tree Construction
Nucleotide sequences from different locations of each species were aligned to determine the location of variations. The cytochrome oxidase subunit (COI) locations were established with the use of the basic local alignment search tool (BLAST), which is also available through the National Center for Biotechnology Information (NCBI) by uploading the sequences and searching for the most comparable reference sequences. The COI sequences of C. burnetii from the GenBank were used for the phylogenetic study. The alignment was produced as molecular evolutionary genetics analysis (MEGA) version 10 (Pennsylvania State University, USA) and FASTA (NCBI, USA) files after being manually altered to remove alignment problems generated by the aligning tool Clustal W. Moreover, a GenBank accession number was issued to each of the acquired nucleotide sequences. Subsequently, the maximum likelihood was applied to analyze and create phylogenetic trees in MEGA version 10 (Pennsylvania State University, USA) (23). One thousand bootstrap samples were used to assess the accuracy of an inferred tree. BioEdit version 7.0.1 (bio informer, Great Bretain) and BLASTn (NCBI, USA) were employed to assess nucleotide diversity through DNA sequence polymorphism analysis.
4. Results
4.1. Nested PCR Enhancement of com1 Gene
Among the 600 milk samples gathered, 74 (12.33%) tested positive for C. burnetii by nested-PCR test, representing a fragment of 438 bp of the com1 gene submitted to the NCBI (accession number OP913466). Additionally, positive results for C. burnetii were found in 38 (12.66%) and 36 (12%) milk samples taken from buffaloes and cattle, respectively. Statistically, the incidence of C. burnetii was insignificant in buffalo and cattle. All the animals selected for milk collection were classified into 3 groups (≤ 6, 7 - 10, and > 10 years old). C. burnetii shedding significantly differed among the age groups of cattle, unlike buffalo groups. According to the results, there were no significant differences between the age groups in buffalos and cattle (Table 2, Figure 3).
| Animal | < 6 (%) | 95% CI | 7 - 10 (%) | 95% CI | > 10 (%) | 95% CI |
|---|---|---|---|---|---|---|
| Buffalo | 10/63 (15.9) | 8.85 - 26.81 | 17/65 (26.15) | 17.02 - 37.95 | 9/70 (12.9) | 6.92 - 22.67 |
| Cattle | 12/66 (18.2) | 10.72 - 29.14 | 14/70 (20) | 12.30 - 30.82 | 12/64 (18.8) | 11.06 - 29.97 |
| Total | 22/129 (17) | 11.54 - 24.47 | 31/135 (23) | 16.67 - 30.74 | 21/134 (15.7) | 10.48 - 22.77 |
The phylogenetic tree of Coxiella burnetii com1 gene sequences was obtained in the present study, and those were deposited in GenBank from different accession numbers. The com1 gene sequences obtained in this study are shown with a bold arrow. The tree was inferred using the neighbor-joining method of MEGA v. 10. Bootstrap values are shown at each branch point. The numbers above branches reflect the bootstrap support of 1000 replicates. All aligned sites containing insertion-deletion or missing data were excluded from the analysis.
4.2. Seasonal Study of Coxiella burnetii Infection in Raw Milk
No positive samples were detected during the winter, while the most contaminated items with C. burnetii were observed in the summer-sampled milk of both cattle and buffaloes (28%, P < 0.05, 95% CI: 21.4 - 35.6%). Based on the results, there was a significant difference (P < 0.05) between seasons in terms of the presence of C. burnetii in the raw milk samples (Table 3).
| Animal | Spring (%) | 95% CI | Summer (%) | 95% CI | Fall (%) | 95% CI | Winter (%) | 95% CI |
|---|---|---|---|---|---|---|---|---|
| Buffalo | 6/50 (12) | 5.62 - 23.8 | 20/50 (40) | 27.61 - 53.82 | 10/50 (20) | 11.24 - 33.04 | 0/50 (0.0) | 0 - 7.13 |
| Cattle | 8/50 (16) | 8.34 - 28.517 | 22/50 (44) | 31.16 - 57.69 | 8/50 (16) | 8.34 - 28.517 | 0/50 (0.0) | 0 - 7.137 |
| Total | 14/100 (14) | 8.53 - 22.14 | 42/100 (42) | 32.8 - 51.797 | 18/100 (18) | 11.7 - 26.677 | 0/100 (0.0) | 0 - 3.7 |
5. Discussion
Query fever is a zoonotic disease that infects all animals. In addition, there is a potential risk of infection in humans through a variety of ways. Individuals involved in animal farming are particularly at risk (20, 24). The results of this study indicated the prevalence of C. burnetii in cow and buffalo milk samples in the Urmia region. Based on the findings, 12.33% of all tested raw milk samples were positive for C. burnetii. The contamination rate of buffalo milk (12.66%) by C. burnetii was greater than that of cattle (12%). The current study results are inconsistent with those of Keshavamurthy et al. in India. They detected C. burnetii in the blood, milk, and vaginal swabs of Indian buffalo by using the trans-PCR and ELISA methods to identify C. burnetii. They further reported C. burnetii in buffalo (8.7%) and cattle (4.3%) (25). This inconsistency can be due to geographical differences, examined sample types, and the management practices of the farms. However, our findings are in line with the results of Khademi et al., which represent a seasonal trend of the presence of Q fever in buffalo and cattle in summer. This consistency is probably due to the similarity of geographical location and climate conditions (26).
Various researchers in several countries reported the prevalence of C. burnetii in cow milk in the range of 4.7 - 53.7% in Switzerland and Japan, respectively. In addition, the prevalence rate differed in different parts of Iran (5.7%, 11%, 20%, 12%, and 26% in Lorestan, Fars, Yazd, Tehran, and East Azerbaijan provinces, respectively). In one of the most recent studies by Khademi et al. (26) in West Azerbaijan province, the prevalence of C. burnetii in cow milk was 14.6%. (8, 9, 26-32). According to reports, the prevalence of C. burnetii was classified into different geographical regions due to various hygiene factors in farms, types of livestock farming, geographical zones, and environments. The higher spread of C. burnetii in cattle in West Azerbaijan (38%), compared with several provinces of Iran, can be explained by the proximity of buffalos and cattle to common grazing areas (33).
Thus, cattle milk is recognized as an essential feature in the epidemiology of Q fever, which may notably influence generic health.
There was a considerable correlation between age and C. burnetii prevalence in this study, specifically for cow milk. This result confirms a previous report indicating that age is a vital risk factor for this condition. The likelihood of a successful outcome also increased by 1.67 times for each extra year of age (34). Raw milk consumption was also demonstrated to vary substantially between regions. Milk contamination with C. burnetii was found to be more prevalent in the southern part of the study area. This follows reports explaining the regional distribution of human cases is similar to the distribution and density of sheep and cattle populations. Thus, shedding the bacterium in buffalo, cattle, and sheep populations may enhance positive specimens (35).
In (36), a seasonal trend of this fever beginning in humans is noted in spring and early summer. It was also observed that the increasing occurrence of Q fever has a close relationship with the lambing season. This finding is in agreement with reports in various European countries, noting that most reported positive cases were during the summer because of the lambing season (4, 21, 24). In the current research, the maximum spread of milk shedding was in summer, which is consistent with recent reports (4, 21, 24). Finally, differences between the results of this study and other studies in Iran and other nations may be due to the persistence of organisms in the environment, climatic conditions, or even the sample size.
5.1. Conclusions
The results of the present study indicate that raw milk from cattle and buffalo is a significant source of Q fever agents. Moreover, age could be a significant risk factor for the presence of C. burnetii in raw milk, which is also associated with seasonal variations. Cattle and buffalo may play a crucial role in the epidemiology of Q fever in the Urmia region, and this should be taken into account for public health purposes.