1. Background
Breast cancer is the first deadly disease in women and has the second highest mortality rate in humans, after heart disease. As such, it has always been a concern for many researchers (1, 2). Finding a method for rapid diagnosis of breast cancer is a major goal of researchers in this field (3, 4). Different diagnostic methods are used for breast cancer, including physical examination, mammography, sonography, magnetic resonance imaging (MRI), bone scan, pathologic sampling, and positron emission tomography (PET) scan (5, 6). In recent years, extensive research has been conducted to diagnose this disease and treat it with various methods. One such method is bioinformatics and molecular docking simulations between drug molecules with cancerous cells or part of these cells, such as membranes, cell receptors, and intestinal organs. In about 20 percent of these patients, the breast cancer cells have human epidermal growth factor receptor 2 (HER2), a protein called HER2-positive cancer. Determining whether a tumor is HER2-positive is clinically important because specific treatments are used for this type of cancer (7).
Human epidermal growth factor receptor 2 (HER2) plays an important role in the development of breast cancer. It has been shown that the excessive expression of HER2 as a prognostic and predictive factor in breast cancer is associated with an increased risk of recurrence and death from breast cancer. On cancerous cells, especially breast cancer, this receptor is overextended and causes the uncontrolled division of the cancerous cells (8).
The LTVSPWY peptide is one of the best-known peptides with high affinity to receptors on the cell surface of HER2 (9, 10). By labeling this peptide with a radioactive material suitable for imaging, the presence of a large HER2 receptor on the cell surface can be hypothesized based on molecular and computational nuclear medicine imaging techniques (11).
Nuclear medicine is a branch of medical and molecular imaging that uses nuclear radioactive properties, such as radioisotopes, to diagnose and treat diseases. The most widely used radioisotope in nuclear medicine is technetium-99 m (Tc-99m). In this method of imaging, radio drugs first enter the patient's body through various methods and are then distributed in the body and accumulate in the particular organ for which the radio drug is made. This accumulation is imaged by gamma-ray cameras (12).
2. Objectives
In contrast to clinical and experimental studies that used radioisotopes, the present study was based on bioinformatics analyses, which is a novel strategy in these criteria (radiopharmaceuticals and nuclear medicine imaging systems) that may lead to the knowledge-based formulation of new drugs in cancer research. This study tried to find a method for imaging of HER2-positive breast cancer tissues via radiopharmaceuticals and nuclear medicine imaging systems.
3. Methods
In the first step, the LTVSPWY peptide structure was received from 2 databases, namely PepBank (http://pepbank.mgh.harvard.edu/interactions/details/56185) and the National Center for Biotechnology Information (NCBI) (National Center for Biotechnology Information) to reduce the probability of errors (Figure 1A).
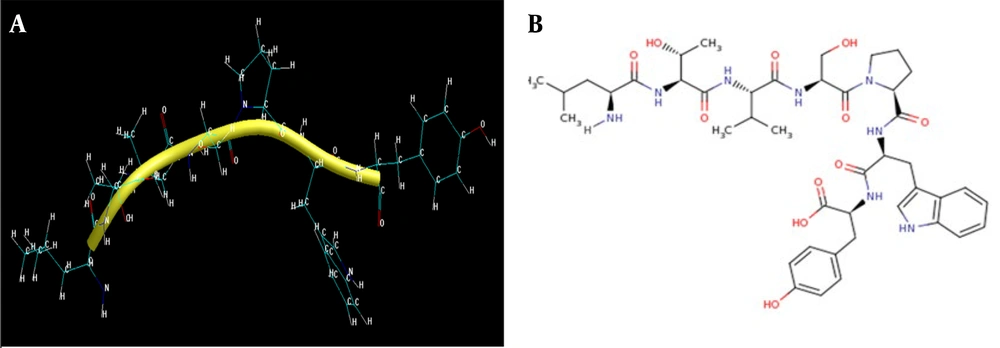
(A) The chemical formula of the peptide's LTVSPWY (https://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?dps=2&uid=2A91); (B) the peptide secondary structure after drawing in HyperChem
This database belongs to the American Harvard University and its medical school, which was launched in the Center for Systems Biology (CSB) and is one of the most important databases for bioinformatics in the United States and the world. This database is known as one of the valid LTVSPWY peptide databases.
One of the most widely used biological information search tools is the Entrez search engine created by the NCBI database. This engine simultaneously searches databases such as PubMed, nucleotide sequences, protein sequences, and protein structures. Currently, the NCBI (http://www.ncbi.nlm.nih.gov) is the most complete and best database containing nucleotide and protein information.
The Entrez search engine uses the close relationship of records in different databases to retrieve and provide the user with biological information on a specific topic from different databases. The structure of the HER2 receptor used was also downloaded from this database and was used in the next steps and in other software. The structure of the peptide, named 2A91 in this database, was extracted from this database (Table 1).
| Variables | Values |
|---|---|
| 2A91: Crystal structure of Erbb2 domains 1-3 | |
| PDB ID | 2A91 |
| MMDB ID | 34415 |
| PDB deposition date | 2005/7/11 |
| Updated in MMDB | 2012/11 |
| Experimental method | X-ray diffraction |
| Resolution | 2.5 Â |
| Source organism | Homo sapiens |
| Address | https://www.ncbi.nlm.nih.gov/Structure/pdb/2A91 |
Abbreviations: HER2, human epidermal growth factor receptor 2; NCBI, national center for biotechnology information.
This peptide is a heptamer; it has 7 amino acids and, therefore, a short sequence. This sequence was entered into the PEPstrMOD (http://osddlinux.osdd.net/rghva/pepstrmod) web server, and the LTVSPWY peptide was registered in this database. Three days later, the results were received from the server. This server has a powerful computing system; it optimizes molecules in terms of energy and structure and also predicts their structure. In addition, this server shows the LTVSPWY peptide energy graph, expressed in kilojoules per mole within the 100 picosecond range, and the root mean square error graph for calculating peptide energy. Peptide hydrophilicity and network charge were calculated using a calculator available on the BACHEM site (https://bachem.com). Moreover, the energy of the peptide, its gradient, and the other components was obtained by HyperChem (http://www.hypercubeusa.com) software (Table 2). Due to the molecular structure obtained from the LTVSPWY peptide after making the necessary corrections, it could be labeled with the radioactive element Tc. HyperChem, a powerful bioinformatics software, was used for peptide labeling. Then, the structure was optimized with molecular mechanics and semi-empirical methods in this software (Table 3).
| Variables | Values |
|---|---|
| Molecular and structural information of the peptide | |
| Molecular weight | 865.00 |
| Isoelectric point | 5.9 |
| Network charge at PH = 7 | 0.0 |
| Average hydrophilicity | - 1.3 |
| The ratio of hydrophilic residues to total residues | 14% |
| Energy and gradient of the optimized peptide | |
| Energy | - 7.146355 kcal/mol |
| Gradient | 0.070165 |
| Converged | Yes (720 cycles 3309 per point) |
| Variables | Values |
|---|---|
| Energy and gradient | |
| Energy | - 27.822974 kcal/mol |
| Gradient | 0.088139 |
| Converged | Yes (0 cycles per point) |
| Total energy and temperature | |
| Time | 1 ps |
| Total energy | 37.5011 kcal/mol |
| Temperature | 157.042 k |
| Structural potential based on the Monte Carlo function | |
| Time | 100 steps |
| Potential | 27.5414 kcal/mol |
| Temperature | 300 k |
Because Tc has different oxidation numbers and a good desire for oxidation, it is best to combine it with lateral oxygen in the desired peptide chain. It seems that the best binding site for a peptide is the oxygen of tyrosine, according to the following reasons: This oxygen has a single bond with the hydrogen on one side; this oxygen is oxygen#24, has a carbon#22 binding peptide structure, and is located on tyrosine (Figure 1B).
In this chain, tyrosine is the second amino acid; therefore, it is closer to one of the two ends of the peptide chain. This position reduces the total energy of the compound and prevents space interference of the chain's electrical charges in the composition. It also facilitates the production of the drug because there is no need to add another composition as a carrier and adhesive bonding to link Tc to the compound, and it connects directly to the main composition. In this study, the new composition was optimized again after adding Tc to the peptide chain.
The ConTEXT version 0.986 program (USA) is widely used in structural modeling, drug design, and molecular dynamics and can be used to add structure to the protein structure or connect protein parts spatially. By means of this program, the protein complex together with the ligand or each separately can be manually edited, which is more accurate. The edits made by this program on our receptor include removing water and heteroatoms, including ligands, cofactors, carbohydrates, and ceyoprotectant.
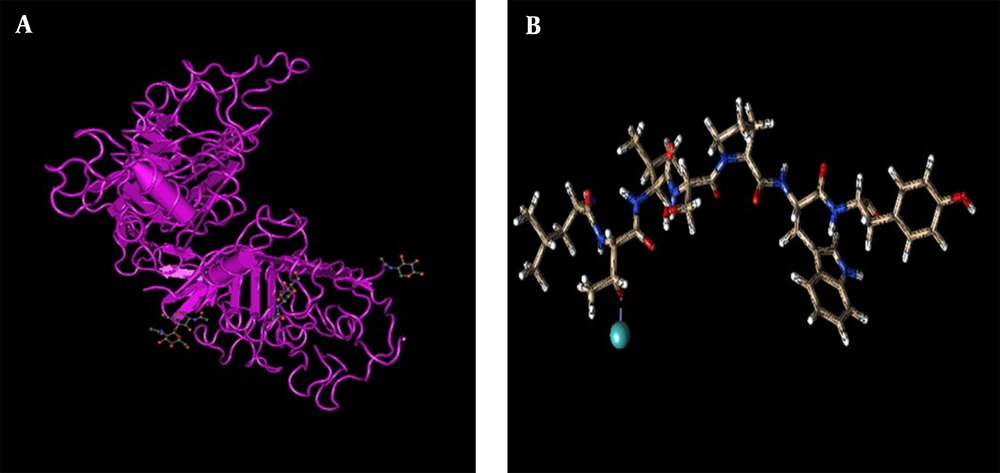
Chimera version 1.10 (California, USA) is a highly extensible program for interactive visualization and analysis of molecular structures and related data, including density maps, submolecular assemblies, sequence alignments, complementation results, pathways, and organizational groups. It can produce high-quality images and animations. Chimera includes full documentation and several tutorials and can be used for free by public, non-profit, and private universities. It is funded by various sources, including Bioinformatics Collections and the US National Institutes of Health. After this step, the following steps were performed on the labeled peptide using the UCSF Chimera (https://www.cgl.ucsf.edu/chimera/) program: Adding the load to the ligand, adding hydrogen to the ligand structure, removing the water molecules (if any) and then, saving the ligand as a PDB file to perform the docking in the HEX program (Figure 2A).
The ligand has a strong affinity for the junction to the HER2 receptor, which is confirmed by optimization software, and the best geometrical and spatial position for the junction was achieved based on calculations. Then, we extracted the HER2 receiver from the databases. First, we searched different databases and reviewed articles and structures to select the appropriate composition and structure of the NCBI database, called 2A91, from other or similar structures (Figure 2B). The structure in question had a very good resolution and was obtained based on X-ray crystallography; 7 years later, it was updated again. It also had major chains of human organisms. It is possible to modify the basic structure of the receptor and protein complex with ligands by removing heterogeneous atoms, including ligands, cofactors, carbohydrates, and water.
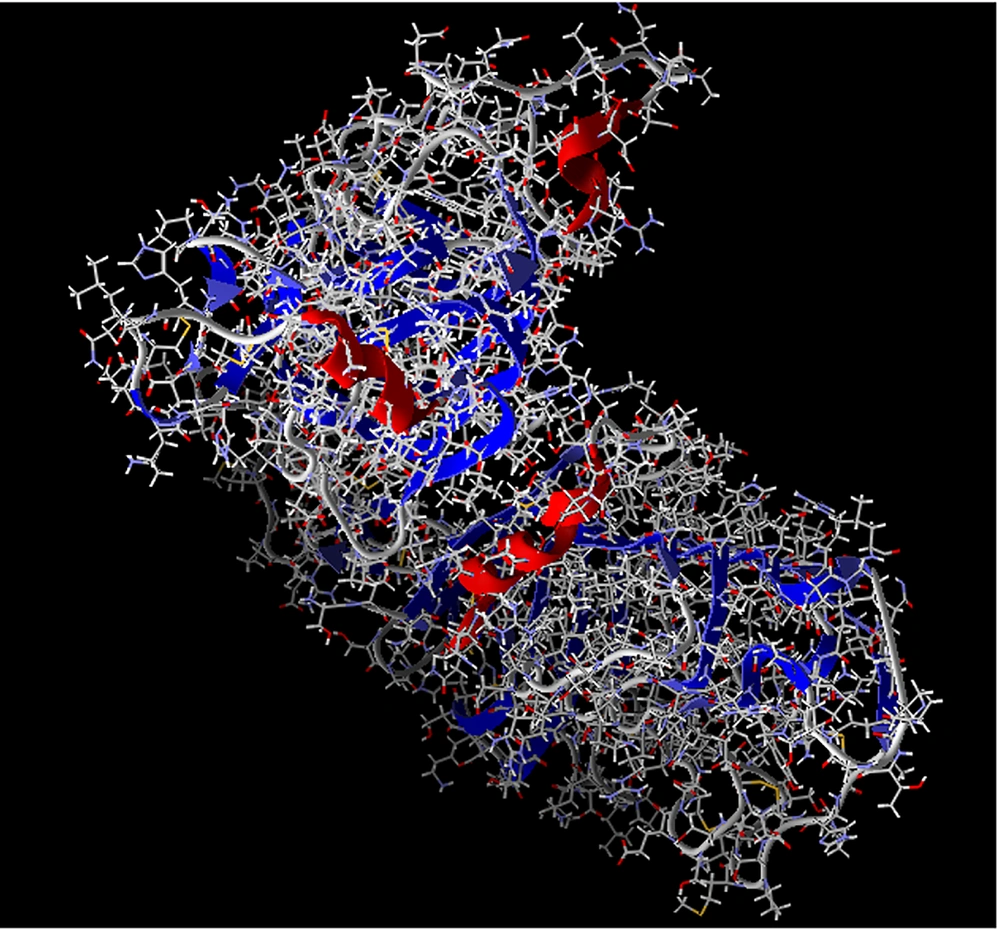
AutoDock (https://autodock.scripps.edu/) is a free collection of docking tools by the SCRIPPS Research Institute. This software allows the ligand to be as flexible as the side chains of macromolecules. The program has a free energy scoring function that calculates the AMBER force field for a large class of protein-ligand complexes based on linear regression tests and scoring functions. The search algorithms in this program include connection simulation, which is a genetic algorithm. Using this program, we prepare our ligand and receptor for docking. Examples of the steps performed by this program on ligands and receptors are adding charge and atomic coordinates to the ligand, which program checks the structure of the ligand in terms of atomic coordinates, charge, and rotation of the atom and makes the necessary changes on them and prepares the receptor for docking. Then, the receptor was inserted into the Molegro Virtual Docker (MVD) program (http://molexus.io/molegro-virtual-docker/), other structural bugs were corrected, and the additional links between the ligand and the receptor that were not detachable in the previous step were removed (Figure 3).
The MVD program is used to study or predict the interaction between ligands and macromolecules, and it works like the AutoCAD program. In this program, in addition to docking, one can trim the docking complex and fix its structural problems to some extent. With this program, it is possible to separate the ligands attached to the receptor structure, which cannot be separated from the main structure with other software. It is also possible to trim and separate the carbohydrates, additional cofactors, and protective carriers from the main composition. Then, the receptor is loaded for the final preparation before docking into the UCSF Chimera software. In this program, the electrical charge and hydrogen are loaded into the receptor structure, and water molecules and other additional substances are extracted from the structure and saved in the PDC file format for the docking step.
The HEX program was selected for docking the labeled ligand to the receptor. After completing the steps of preparing the ligand and receptor and storing them in the appropriate file format, they were entered into the HEX (https://hex.loria.fr/) software, and the conditions were set for docking to select the largest possible grade. The conditions were set for the highest possible sensitivity and docking. The HEX program is one of the most widely used free docking software, which also has a web server for which there are more settings for docking. It is useful for those who do not have access to a powerful computer. This program uses polar-spherical Fourier correlation (SPF) for docking calculations.
Docking procedures with this software are as follows: Trimming the structure of the receptor that includes removing water, ligands, and extra chains of the desired receptor, adding charge and hydrogen to both receptor and ligand structures, performing docking, recording the docking results and filing it for review in the next steps. In this program, the larger molecule is always considered the receptor, and the smaller molecule is considered the ligand.
After the docking step, the resulting data were entered into the LIGPLOT + program (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/). LIGPLOT + identifies the position of spatial ligands and determines which amino acid is located near it. It also determines the role or position of these amino acids in the structure. After selecting the right ligand and applying some restrictions on the geometrical parameters of the structures, the most sensitive grade of the program was chosen, and docking was performed such that the rate of errors was kept as low as possible. Affinity calculations were also performed based on the results to validate the docked conformations, and adequate convergence was revealed.
4. Results
4.1. Preparation of the Receptor and Ligand
Based on the analyses, it was deduced that the finest structures for both the ligand and receptor were achieved, and the best correlations according to the scoring algorithms were extracted; this means that the most convergent conformations of complexes were produced by the screening calculations.
4.2. Docking Success
Increasing the binding energy of docking with a technetium-labeled peptide in this study compared to a nonlabeled sample is a sign of the success of docking and its proper steps (Table 4).
| Variables | Values |
|---|---|
| The Docking End Energy of the Ligand is Not Labeled by the Receptor | |
| RMS | - 1:00 |
| Bumps | 0 |
| H-Bonds | - 1 |
| Models | 0:0 |
| Solution | 1 |
| Cluster | 1 |
| Vclash | 0.00 |
| Vshape | 0.00 |
| Eair | 0:00 |
| Eforce | - 43.23 |
| Eshape | - 138.59 |
| Etotal | - 130.43 |
| The Docking End Energy of the Ligand Labeled by the Receptor | |
| RMS | - 1:00 |
| Bumps | 0 |
| H-Bonds | - 1 |
| Models | 0:0 |
| Solution | 1 |
| Cluster | 1 |
| Vclash | 0.00 |
| Vshape | 0.00 |
| Eair | 0:00 |
| Eforce | - 48.12 |
| Eshape | - 165.46 |
| Etotal | - 213.58 |
4.3. Evaluation of Connection Results and Connection Success
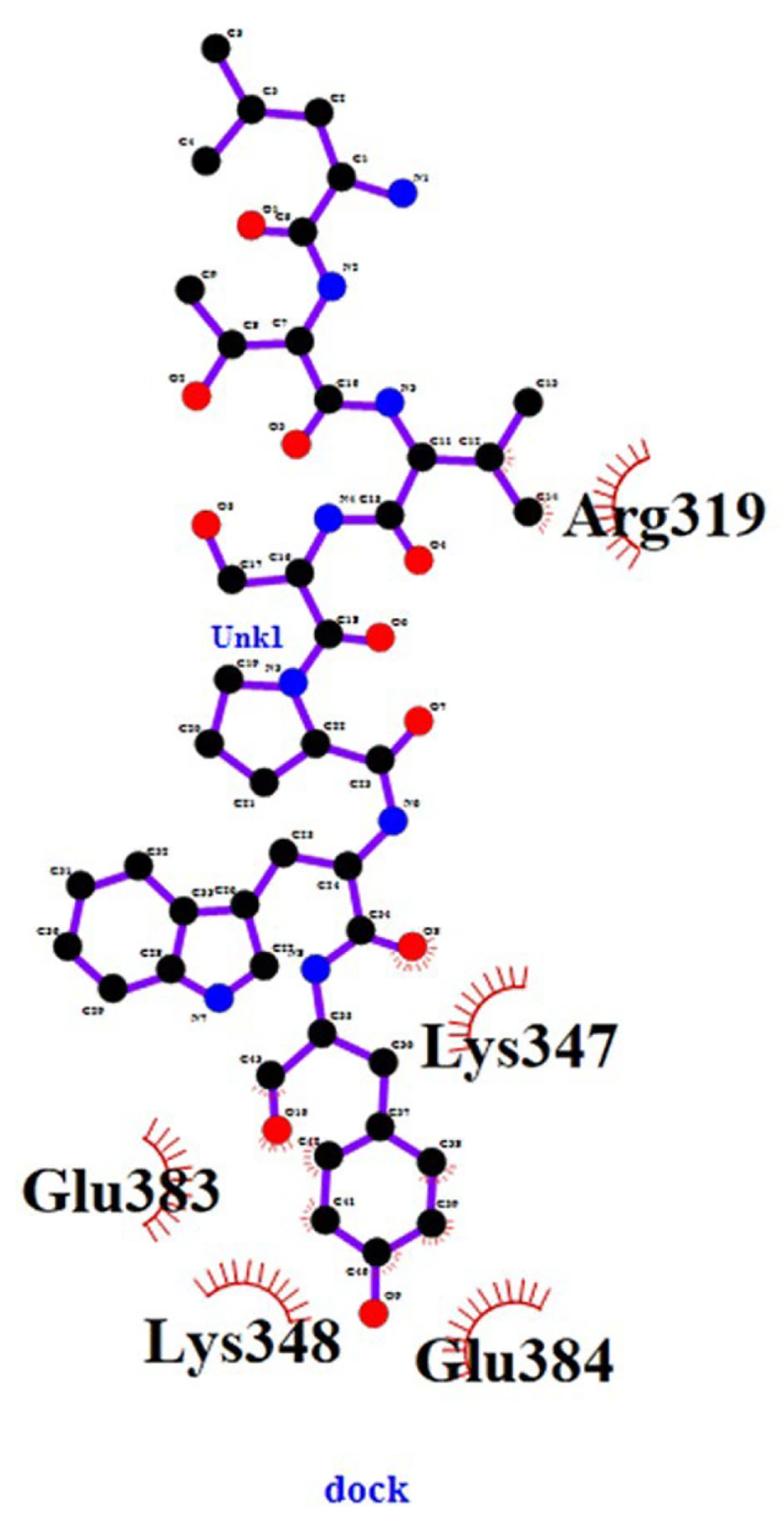
In the analysis of data after docking with the LIGPLOT + software, we observed that the ligand was connected to the HER2 receptor from 5 major points, indicating a good and proper bond between the ligand and the receptor. These amino acids were arginine, lysine, glutamic acid, lysine, and glutamic acid, showing that the ligand was attached to the receptor by a strong bond at the beginning of the chain and 4 other strong links at the end of the chain, with 2 links on one of the rings.
4.4. Checking the Location and Connection Positions
At the beginning of the ligand chain, arginine is bonded to two carbon 12 and 14 ligands, and this connection in more than one point represents a solid connection at the beginning of the ligand chain with the receptor; at the end of the chain, these connections are more and much better. At the end of the chain, a lysine amino acid is bonded from the receptor to ligand oxygen # 8. It is clear that oxygen is a highly electronegative atom active in chemical reactions and creates a strong bond. Moreover, a glutamic acid receptor is linked to three carbon atoms 41, 42, and 43 ligands. Another lysine from the receptor with oxygen number 10 of the ligand and another glutamic acid receptor is also linked to 39 and 40 carbon-chain ligands. Of the 10 atoms connecting the ligand to the receptor, which includes 8 carbon atoms and 2 oxygen atoms, 2 carbon atoms are at the beginning of the chain, and 8 atoms containing 6 carbon atoms and 2 oxygen atoms are at the end of the chain. Therefore, the end of the chain has a stronger connection than its beginning. These results confirm the success of the molecular docking and express the good linkage of the ligand to the receptor after its evaluation. The connection between two regions at the beginning and end of the ligand chain to the receptor is important because the ligand chain is more stable after binding to the receptor and reduces the probability of separation from the receptor. It also helps stabilize the ligand chain. All the cases described indicate the successful labeling of the ligand and its binding to the receptor, which has been well-illustrated in the simulation and docking of the ligand with the receptor (Figure 4).
The amino acids present are all hydrophilic, which makes them work better in the labeled peptide. The polarity of glutamic acid is also very important in this connection because, despite its positive charge and the possibility of its involvement in the connection, its polarity also strengthens the possibility of the formation of polar bonds between the ligand and the receptor, which is a very strong connection. This phenomenon is expected. The total number of atoms that have participated in binding the ligand to the receptor is 10 atoms. There are 8 carbon atoms and 2 oxygen atoms, 2 carbon atoms at the top of the chain, and 8 atoms (including 6 carbon atoms and 2 oxygen atoms) at the bottom of the chain, which are connected to the receptor.
Connecting the ligand to the receptor from the top and bottom regions is important in the sense that the ligand chain is more stable after binding to the receptor, and this reduces the possibility of its separation from the receptor. It will also help the stability and stability of the ligand chain. Since the binding of arginine at the top of the chain to carbons 12 and 14 of the ligand chain is close to our technetium (which is close to the oxygen attached to carbon number 22 of the ligand chain), it increases the probability of receptor labeling by our labeled ligand.
5. Discussion
Compared to the research of Sobhnoo et al., it can be seen that in the bioinformatics method, labeling with the addition of a ligand has a direct relationship with its original structure. Moreover, labeling can be performed without changing the structure of the peptide, and there is no problem with the heavy peptide chain due to the addition of additional ligands to the structure, which results in instability in labeling stabilization (13).
Docking results obtained in this research were compared with the research by Jie et al. The results are consistent in the diagnosis and identification of the tumor by the desired peptide (14). In this research, the same peptide was used, and magnetic nanoparticles modified with LTVSPWY-PEG-CS were prepared using the solvent diffusion method and used for MRI imaging. The results proved tumor detection and identification by this peptide. This can be a good justification for using this peptide in other diagnostic imaging methods, such as nuclear medicine, which has a more physiological aspect than MRI and opens the way for more research in this field.
This research is also consistent with the one conducted by Zhang et al., who used small peptides for targeted drug delivery. In this study, this small peptide has a special receptor to detect and deliver a diagnostic drug (15).
Since arginine amino acid is a positively charged amino acid, its positive charge is probably the main cause of chain bonding, and this helps to strengthen docking (16).
The amino acid lysine is also a positively charged amino acid, which probably plays a role in this interaction and binding (17).
Glutamic acid is an amino acid with a negative charge, and the connection of 3 carbon atoms with this amino acid has probably caused the excessive strength of these bonds. Pulling the negative charge of this amino acid towards itself causes the spread of excess charges, reduces other reactions, and strengthens the connection. By reducing the charge, the absence of excess charge at the connection site, and the interference and defense of charges of the same name, it prevents the rupture of the connection and sequence of peptides (18). Shadidi and Sioud also found that the peptide was widely implicated in breast cancer cells and could be used to target them. In their study, the peptide was labeled with fluorescence, and the results demonstrated the success of peptide binding to the HER2 receptor in breast cancer cells, which is confirmed by the results of the present study (19). Wang et al. showed that the peptide can be used for drug delivery, the control of the growth rate of cancer cells, and their treatment. The results of this study were also positive; ligand binding to the receptor was successful, and excessive expression of the receptor was inhibited. This study demonstrated the successful ligand and receptor binding and confirmed our research results (20).
According to the study by Orban et al., this peptide was once again used to check the binding to the ErbB2 receptor; it was also found that in various cancerous cells with high expression of ErbB2, this peptide had an acceptable connection, and had successful results in an animal sample (21). In 2003, Shadidi and Sioud used a peptide linked to another ligand peptide against cancer cells. The outcomes were successful, indicating the successful binding of the peptide to the receptor and the use of the peptide for drug delivery (22).
In recent research by Khodadust et al., This peptide was used to deliver the Tc-labeled drug to tumors with overexpression of HER2. The results were successful and confirmed the findings of our research (23). Besides, in our research compared without marking, the increase in docking binding energy of the technetium-labeled sample was a sign of successful docking.
Furthermore, according to the researcher's experiences with nuclear medicine and the drugs used in medical sciences and nuclear medicine, the marking steps presented in this study were very similar to the marking of red and white blood cells in medical scans. In any field of medical and pharmaceutical sciences, it is more acceptable to use previous methods that have been proven, and their process is more specific. In nuclear medicine, due to the presence of radioactive materials, there are many limitations in conducting research, and the use of unknown elements seems very risky, especially since the separation of the radioactive part from its main chain has its own consequences. However, using technetium as a radioactive tracer, which is the most widely used element in nuclear medicine and one of the least harmful radiopharmaceuticals, is the most economical and reliable choice.
Docking with bioinformatics methods is actually geometrical description, molecular dynamics analysis methods, and the use of chemical properties required for ligand binding with its receptor. The purpose of molecular design in bioinformatics is to create new techniques to reduce the overall costs of the discovery and development of a new drug; thus, by considering the receptor model and identifying potential candidates to be selected as ligands, an effective method can minimize the rate of failures in experimental efforts. Most of the pharmaceutical projects that fail in Iran and other countries are often due to not considering bioinformatic and pharmacophoric models in the design of drugs that are optimized without any assumptions of screening ligands and receptors. Currently, worldwide, experimental ligand and receptor models have serious weaknesses and failures to identify the most favorable ligands in the design of drugs since bioinformatics models and calculations can easily increase costs. Bioinformatics methods and calculations have proven that, at a very precise level, they can increase the success rate in the production and design of various types of drugs.
5.1. Conclusions
Considering the results of this study and comparing them with other studies, most of which were clinical and in vitro, it seems that the LTVSPWY ligand labeled with Tc radioactive element for binding to the HER2 receptor in patients with breast cancer has been successful, with the excessive expression of the HER2 receptor by bioinformatics and molecular docking techniques. Therefore, the effective and successful outcomes of this study can be used in clinical and laboratory research.