1. Background
Colorectal cancer (CRC), one of the most common malignancies worldwide, has a poor prognosis and extensive metastatic dissemination. Radiotherapy is a crucial procedure for CRC therapy, but its use is limited by radioresistance. In addition, surgery is an option for advanced CRC, which is frequently followed by further chemotherapy (1, 2). Although CRC screening can reduce the incidence and mortality rate, it is not usually performed. On the other hand, blood tests are the preferred fundamental way of detection. As a result, more markers must be discovered (3). Predictive biomarkers provide valuable details for directing therapeutic decisions in patients who are favorable for the biomarker compared to those who are negative (4). Despite improvements in traditional screening methods and the development of promising medications, the survival rates for CRC remain low. The increased incidence is ascribed to illnesses, poor diet, and obesity (5).
Long non-coding RNAs (lncRNAs) that are not translated to proteins have critical roles in gene expression regulation, epigenetic modifications, cell growth, and differentiation (6). Furthermore, lncRNAs are dysregulated in several malignancies, and they are regarded as important prognostic and diagnostic biomarkers because of their distinct expression patterns and high tissue- and cell-specificity (7). In addition, recent research has demonstrated that lncRNAs may be involved in migration, metastasis, chemoresistance, epithelial-to-mesenchymal transition (EMT), and proliferation of CRC (8-12).
According to research by Liu et al., VLDLR-AS1 regulated the expression of genes related to lipid loss in cancer cachexia, responsible for 20% of all cancer-related deaths. Further investigation discovered that genes associated with VLDLR-AS1 were implicated in EMT, the Wnt signaling pathway, and small GTPase-mediated signal transduction (13). Chen et al. reported that esophageal cancer cells had increased levels of VLDLR-AS1, an exosomal lncRNA, that led to chemoresistance. Furthermore, they revealed that the extracellular vesicles (EVs) generated by drug-resistant cells might upregulate the expression of ABCG2 and, hence, affect drug resistance related to the VLDLR-AS1 carried by EVs (14). By comprehensive RNA-seq data analysis in patients with thymomas, Ji et al. discovered 65 differently expressed (DE) lncRNAs. VLDLR-AS1 was overexpressed and validated by The Cancer Genome Atlas (TCGA) (15).
2. Objectives
In the current study, VLDLR-AS1 expression levels were evaluated in the CRC tissues compared to healthy tumor margins for the first time. The biomarker potency of VLDLR-AS1 for discriminating tumoral and non-tumoral CRC tissues was also investigated.
3. Methods
3.1. CRC Tissue Sampling
Ninety-four paired samples of cancerous and non-cancerous adjacent tissues were collected from patients with CRC referred to the Noor-e Nejat Hospital in Tabriz, Iran. Samples were immediately placed in liquid nitrogen, transferred to the laboratory, and stored at -80°C. The Ethical Committee of the University of Tabriz approved this study. Written informed consent was obtained from all patients. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki, as reflected in a prior approval by the institution's human research committee.
3.2. RNA Extraction, cDNA Synthesis, and qRT-PCR
Total RNA was extracted from tissues using TRIZOL reagent (Invitrogen, Massachusetts, USA) based on the manufacturer's guidelines. Thermo Fisher Scientific NanoDrop (2000, CA, USA) was used to assess the purity and quantity of the isolated RNAs. The integrity of RNA molecules was evaluated using 2% gel electrophoresis. DNase I (Gene All, Korea) treatment was performed to remove residual DNA contaminations. For cDNA synthesis, PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa, Kusatsu, Japan) was utilized. Gene-specific primers were designed by the Gene Runner software, and the BLAST program evaluated their specificity. Sequences of the primers were VLDLR-AS1 forward: 5’-GAATCGAAGCGCCCCTATCA-3’, reverse: 5’-CAGTGAGGCAAGCTGGTTCT-3’, U6 forward: 5’-CTCGCTTCGGCAGCACAT-3’, U6 reverse: 5’-GGAACGCTTCACGAATTTGC-3’. The U6 was used as internal control. Expression of VLDLR-AS1 and U6 was assessed using StepOnePlusTM Real-Time PCR System (Applied Biosystems). The reaction mixture, including SYBER Green Master Mix (Amplicon Company) 5 µL, forward primer 0.12 µL, reverse primer 0.12 µL, cDNA (diluted 1:100) 4 µL, and ddH2O 0.76 µL, in a final volume of 10 µL was used for amplification. The cycling program was 10 min at 95°C, 45 cycles of 30 sec at 95°C, 30 sec at 58°C, 30 sec at 72°C, followed by 5 min at 72°C. The reactions were duplicated, and the results were normalized to U6. The fold change of VLDLR-AS1 was calculated using the 2-ΔΔCT method.
3.3. Statistical Analysis
The SPSS version 27 and GraphPad version 9 were used for data analysis. The normality of the data was examined using Kolmogorov-Smirnov test. The students' t-test compared the data between two groups. The Mann-Whitney and one-way ANOVA tests were applied to study the association of clinicopathological features with the expression of VLDLR-AS1. The receiver operating characteristic (ROC) curve was utilized to evaluate the biomarker potential of VLDLR-AS1. P-value < 0.05 was considered statistically significant.
4. Results
4.1. Clinicopathological Characteristics of Study Subjects
A total of 188 tissue samples, including 94 tumor and 94 corresponding tumor marginal samples from 94 CRC patients, were studied. The clinicopathological attributes of the patients are described in Table 1; 16 were female, and 78 were male. Concerning age dispersion, 8 cases were ≤55 years old, and 86 were >55 years old.
| Clinical Parameters | No. (%) | P-Value |
|---|---|---|
| Age | 0.22 | |
| >55 | 86 (91) | |
| ≤55 | 8 (9) | |
| Gender | 0.46 | |
| Female | 16 (17) | |
| Male | 78 (83) | |
| Location | 0.72 | |
| Colon | 70 (74) | |
| Rectum | 24 (26) | |
| Invasion | 0.25 | |
| Yes | 80 (85) | |
| No | 14 (15) | |
| Lymph | 0.33 | |
| Yes | 34 (36) | |
| No | 60 (64) | |
| Smoking | 0.42 | |
| Yes | 70 (74) | |
| No | 24 (26) | |
| TNM | 0.24 | |
| I/II | 58 (62) | |
| III/IV | 36 (38) |
4.2. VLDLR-AS1 Downregulation in CRC Tissue Samples
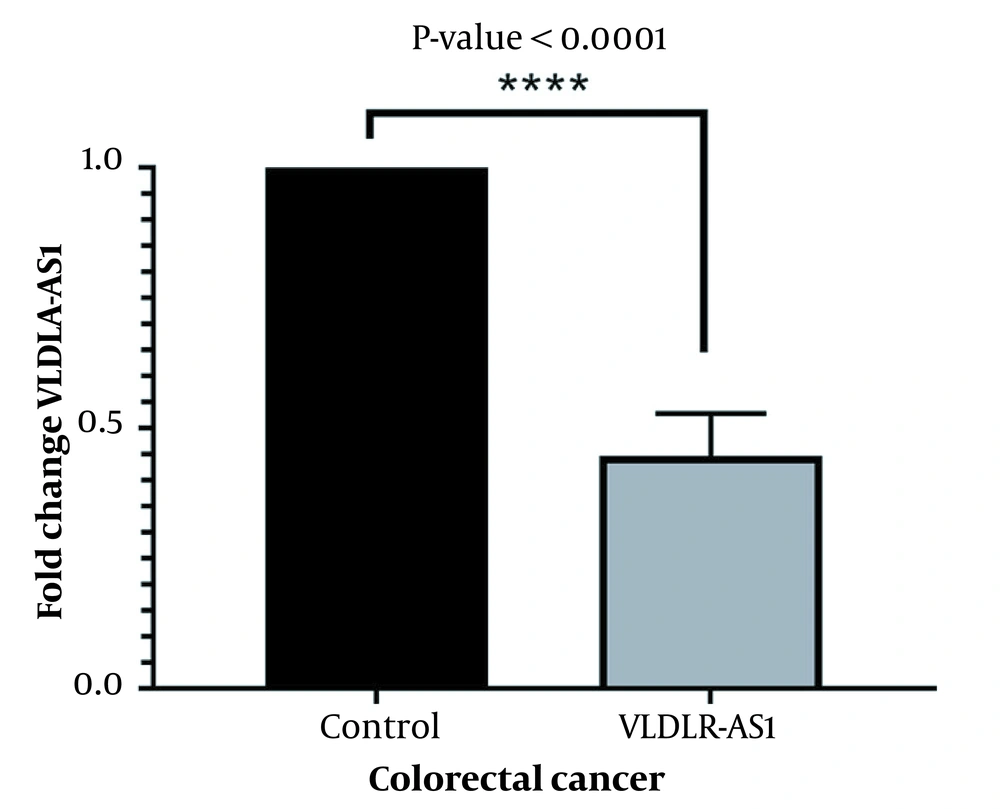
The fold change of VLDLR-AS1 lncRNA was assessed by qRT-PCR. The results showed that the downregulation of VLDLR-AS1 in tumor samples had a 0.44 ± 0.08 fold change compared to the marginal samples (Figure 1, P < 0.0001).
4.3. Correlation of VLDLR-AS1 Expression with Clinicopathological Features
The correlation between VLDLR-AS1 expression levels and clinicopathological characteristics of CRC patients, including age, gender, tumor location, invasion, lymph, smoking habits, and TNM, were analyzed. The results showed no significant correlation between each feature and VLDLR-AS1 expression.
4.4. Biomarker Potential of VLDLR-AS1
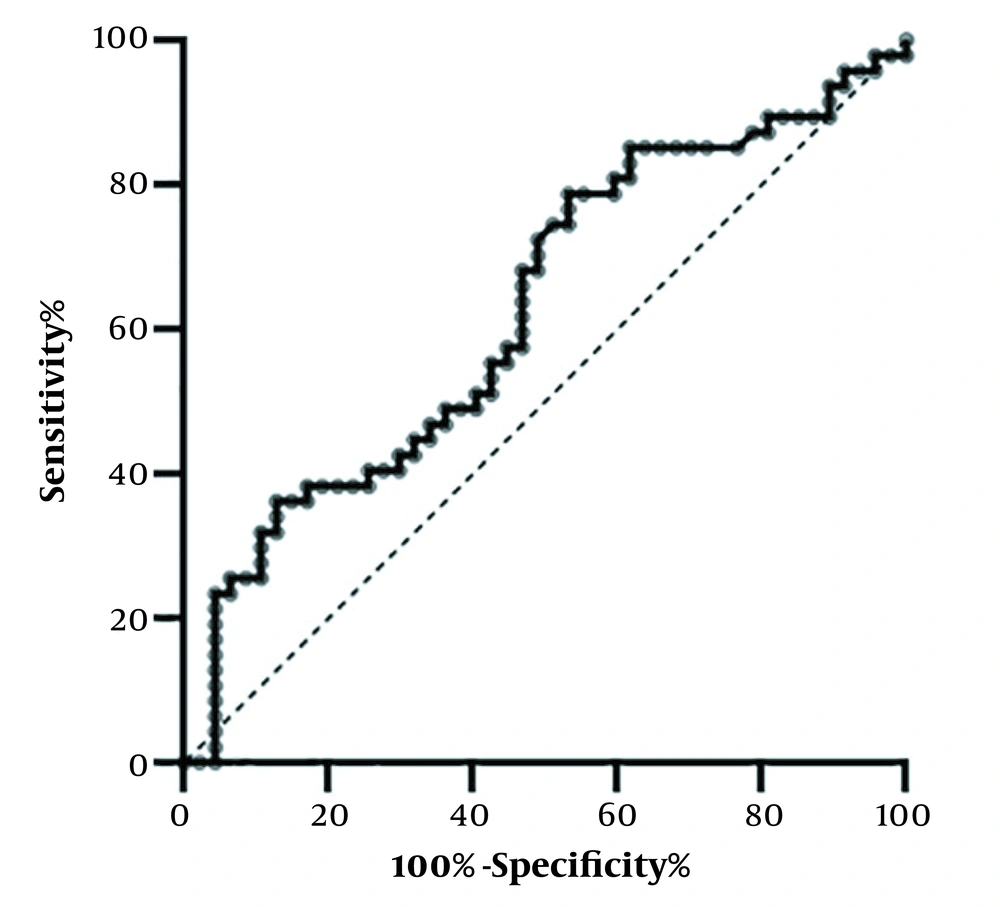
The ROC curve analysis was used to evaluate the biomarker potency of VLDLR-AS1 expression levels for CRC (Figure 2). Table 2 shows that VLDLR-AS1 expression can distinguish tumor and non-tumor samples with a sensitivity and specificity of 72.34% and 51.06%, respectively (P = 0.03, AUC = 0.6274). In addition, the cutoff value (0.0028), standard error (0.0579), and 95% confidence interval (0.5139 - 0.7410) were calculated for the two groups. The results showed that VLDLR-AS1 could be considered a potential diagnostic biomarker.
| ROC Curve Data | AUC | Sensitivity | Specificity | Cutoff Value | Std. Error | P-Value | 95% CI |
|---|---|---|---|---|---|---|---|
| VLDLR-AS1 | 0.6274 | 72.34% | 51.06% | 0.0028 | 0.0579 | 0.03a | 0.5139 - 0.7410 |
a P < 0.05
5. Discussion
CRC is common worldwide, and younger people are being affected more frequently. Further investigations must be conducted to prevent new cases and fatalities (16). The primary therapeutical approach for metastatic CRC is chemotherapy, either alone or in combination with target therapy. It is challenging to achieve advancements in traditional approaches regarding the side effects of chemotherapies and molecular features of the tumor cells (17). As a result, lncRNAs have been identified as beneficial indicators for cancer onset and cell growth (9, 18). Comprehending the role of lncRNAs in cancer biology could be helpful for the treatment, prognosis, and diagnosis of cancer (7, 19, 20). In the current study, for the first time, we investigated VLDLR-AS1 expression levels in CRC and compared its expression levels between tumoral and non-tumoral margin tissue samples.
We found that CRC tissues had significantly reduced levels of VLDLR-AS1 expression compared to the marginal tissues. The results of Zheng et al. were consistent with our research. They indicated that in ovarian cancer (OC), VLDLR-AS1 is enriched in ovarian steroidogenesis or the GnRH signaling pathway. Furthermore, the downregulation of VLDLR-AS1 was related to poor survival and appeared to be an oncogene in OC cellular function (21). In addition, Wu et al. demonstrated that as a protective gene in OC development, the downregulation of VLDLR-AS1 was related to poor survival (22).
In contrast to our findings, Takahashi et al. revealed that VLDLR-AS1 was significantly upregulated in hepatocellular carcinoma. The EVs from tumor cells containing VLDLR-AS1 contributed to cellular stress responses (23). Moreover, malignant hepatocytes had increased VLDLR-AS1 and lincRNA-ROR (linc-ROR) levels, enriched explicitly inside EV and subsequently stimulated by TGFβ to increase EV release. Altogether, these findings provide evidence for the existence of TGFβ-mediated mechanisms for the precise enrichment of these lncRNAs inside EV and their possible roles in intercellular signaling affected by TGF as prospective drug resistance regulators (24).
Moreover, we found that VLDLR-AS1 could be regarded as a possible biomarker, in accordance with the research conducted by Yue et al. These authors also showed that according to survival analysis, patients with lung adenocarcinomas had better prognoses and overall survival rates once VLDLR-AS1 expression levels were raised. A database called TCGA_LUAD was used to verify the mentioned result (25). In addition, Gong et al. revealed that the overexpression of LINC00324, AFAP1-AS1, and VLDLR-AS1 lncRNAs was associated with the enhanced proliferation of thymomas. Furthermore, the VLDLR-AS1 expression profile was linked to relapse-free survival of patients, which could be helpful as a prognostic biomarker (26). In addition, using TCGA data, Ye et al. showed that 76 DE lncRNAs, including VLDLR-AS1, were found in gastric cancer. Further investigation revealed that VLDLR-AS1 and the other 8 hub lncRNAs might play a role in the MAPK signaling pathway. Lower patient overall survival was linked to greater levels of VLDLR-AS1 (27). Overall, TGFβ (24), MAPK (27), and Wnt (13) signaling pathways leading to EMT and chemoresistance (13) may all be altered by VLDLR-AS1 via EVs released from cancer cells. Moreover, in this study, there was no significant association between the expression levels of VLDLR-AS1 and the clinicopathological features of patients with CRC. More research is required because VLDLR-AS1 lncRNA and its critical roles have not been sufficiently studied.
5.1. Conclusions
Downregulation of VLDLR-AS1 lncRNA in CRC tumor tissue was indicated compared to healthy tumor margin tissue. In addition, VLDLR-AS1 might be considered a biomarker for diagnosing CRC progression.