1. Background
In contemporary times, according to reports from the World Health Organization (WHO), cancer stands as the predominant cause of mortality on a global scale (1). Metastases, ultimately manifesting in the majority of cancer instances, assume a pivotal role as a critical prognostic indicator, portending an unfavorable clinical outcome (2). Despite the intricate nature of metastasis in cancer cells, recent strides in molecular biomarkers have significantly advanced the realms of cancer prediction and therapeutic interventions (3).
Hypoxia, a prominent hallmark of solid tumors, precipitates heightened resistance to treatment in patients and augments the progression of neoplastic growth. The hypoxic milieu elicits a molecular response in both normal and neoplastic cellular populations, attributed to the compromised supply of oxygen and nutrients within the local vascular environment. This response triggers the activation of a pivotal transcription factor denoted as hypoxia-inducible factor (HIF) (4). Hypoxia-inducible factor 1α (HIF1α), serving as a master transcriptional regulator, engages in heterodimerization with the HIF1β (also recognized as ARNT) subunit. This complex recognizes hypoxia response elements (HREs) in the genome, binding to the consensus sequence G/ACGTG (5).
The consequences of hypoxia encompass not only the promotion of tumor progression through heightened angiogenesis (6) but also the induction of cell death, as evidenced by the emergence of a central necrotic zone within tumors (7). Furthermore, a corpus of scientific inquiry has unveiled the propensity of hypoxia to instigate and sustain genetic instability and a mutator phenotype. These processes contribute to resistance against apoptosis and a reduction in capacity for deoxyribonucleic acid (DNA) repair. Of particular note, the dysregulation of genes implicated in the occurrence of double-strand breaks (DSBs) in genomic DNA, a process characterized by high carcinogenic potential, has been elucidated in various studies following chronic exposure to hypoxic conditions (8-10).
Homologous recombination (HR) and non-homologous recombination (i.e., end-joining or NHEJ) represent the principal pathways governing the repair of DNA DSBs (11). Within the non-homologous end joining (NHEJ) pathway, XRCC4 assumes a pivotal role by directly interacting with Ku70/Ku80. Noteworthy studies suggest that impeding XRCC4 function might harbor therapeutic potential, enhancing the radio-sensitivity of cancer cells (12-14). Additionally, XRCC7 (PRKDC) contributes to NHEJ by recognizing and mending DNA DSBs (15), and its association with various cancers has been documented in prior research (16-18).
Given the prevalence of DSBs in cancerous cells, potentially linked to hypoxia, this study pioneers an assessment of the expression levels of two crucial NHEJ-related genes, XRCC4 and XRCC7, in HIF1α-knockdown cells under hypoxia induction. This exploration seeks to unravel novel insights into the intricate interplay between hypoxia, HIF1α, and the NHEJ repair pathway, shedding light on potential avenues for therapeutic intervention in cancer treatment.
2. Methods
2.1. Cell Culture
To explore the correlation between HIF1α and XRCC4/XRCC7, two cell lines—human embryonic kidney (HEK293) and HeLa (Pasteur Institute of Iran)—were selected based on their favorable expression levels of the candidate genes. Cell cultures were maintained in Dulbecco's modified eagle medium (DMEM) from Inoclon (Iran) supplemented with 10% fetal bovine serum (Gibco), streptomycin (100 µg/mL), and penicillin (100 U/mL, Gibco). The cells were incubated under standardized conditions of 5% CO2 at 37°C and underwent passaging through trypsinization for experimental continuity.
2.2. Small Interfering Ribonucleic Acid Design and Treatment
For the downregulation of HIF1α gene expression, a specifically designed small interfering ribonucleic acid (siRNA) was generated utilizing the “Oligowalk” online software (19). The anti-sense sequence of the HIF1α siRNA was 5’-FAM-ACATTCACGTATATGATACCATT-3’. The transfection of the FAM-labeled siRNA targeting HIF1α and a control siRNA was accomplished using Lipofectamine 2000 (Invitrogen), following the manufacturer’s guidelines. The efficacy of transfection was assessed through fluorescence detection using microscopy.
2.3. Cell Viability Assay
To assess the impact of the designed siRNA on cell viability, a colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Roche, Germany) was employed. The experiments were conducted over two distinct time intervals (24 and 48 hours). In brief, cells were seeded at a density of 103 cells/well in 96-well culture plates and transfected with siRNA for the specified duration. Subsequently, 10 µL of a 5 mg/mL MTT solution was added to each well, achieving a final concentration of 0.5 mg/mL. The plates were incubated for an additional 4 hours, followed by the removal of the medium and the addition of 100 µL of solubilizing buffer (Roche, Germany) into each well. Microplate readings were performed at 540 nm. Cell viability was quantified as a percentage relative to the control culture value.
2.4. Cell Cycle Analysis
For cell cycle analysis, propidium iodide (PI) staining coupled with flow cytometry was employed. In this procedure, 5 × 104 cells were initially seeded in 24-well plates for 24 hours, followed by transfection with siRNA for a subsequent 48 hours. Subsequently, the cells were trypsinized, phosphate-buffered saline (PBS)-washed, and fixed with 70% ethanol overnight at 4°C. The harvested cells were then re-suspended in PBS containing 100 μg/mL RNase A and 100 μg/mL PI (Invitrogen, Leiden, The Netherlands) for a 30-minute incubation period. Flow cytometric measurements were conducted using a Partec flow cytometer (Münster, Germany).
2.5. RT-qPCR
To evaluate the gene expression, total RNA was extracted using the RiboEX solution (GeneAll, Korea), according to the manufacturer's instructions. Oligo dT and Random Hexamer primers were used for complementary DNA (cDNA) synthesis, which was carried out by 3 μg of extracted total RNA using the 2-step RT-PCR kit (Vivantis Technologies, Malaysia) in a total volume of 20 μL reaction mixture. RT-PCR was used to evaluate the down-regulation of messenger RNA (mRNA) expression of HIF1α in 24, 48, and 72 hours after transfection. Real-time PCR was performed by StepOne Applied Biosystems (Applied Biosystems, USA). The mRNA expression levels were examined for HIF1α by (forward: 5’-GCTTGCTCATCAGTTGCCACTTCC-3’ reverse: 5’-TTTCTCTCATTTCCTCATGGTCAC-3’), XRCC4 (forward: 5’-GCTCCTCAGGAGAATCAGCTTC-3’ reverse: 5’-TACGGTAATAGCGGCTGCTGAC-3’) and XRCC7 (forward: 5’-GCTCTGATATGCATCAGCCACTGG-3’ reverse: 5’-GGAGGGCTCCTTGACAAACACATC-3’) mRNAs were normalized to ACTB (forward: 5’-AGCCTTCCTTCCTGGGCATGG-3’, reverse: 5’-AGCACTGTGTTGGCGTACAGGTC-3’) as an internal control gene. All of the samples were tested at least in duplicate, and the specificity of qPCR reactions was verified by a single band after agarose gel electrophoresis and melting curve analysis.
2.6. In Silico Study
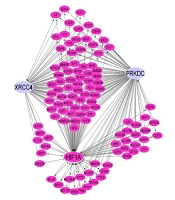
The eukaryotic promoter database (EPD) was used to evaluate the promoter of XRCC4 and XRCC7 genes for HIF1α binding site. To understand the organization and complexity of XRCC4 and XRCC7 gene regulation, we used a gene regulatory network (GRN), which indicates regulatory interactions between transcription factors (TFs) and their target genes. To prepare this network, the transcriptional regulatory relationships unraveled by sentence-based test TRRUST-V2 mining, human transcription factors targets (hTFtarget), regulatory network repository (RegNetwork), and PAZAR databases were queried for the common TFs list for XRCC4 and XRCC7 (or PRKDC) which are regulated by HIF1α. The information was visualized as a network using Cytoscape_v3.8.0 software.
2.7. Statistical Analysis
Statistical analysis was performed by one-way analysis of variance (ANOVA) test followed by Fisher's protected least significant difference posttest for multiple comparisons using the GraphPad Prism software (GraphPad Software, Inc., San Diego, CA, USA). All results were expressed as mean ± standard deviation (SD) of three identical experiments, each performed at least in three replicates. The significance level was considered P < 0.05.
3. Results
3.1. Impact of HIF1α Downregulation on Cell Viability in HeLa and HEK293 Cells
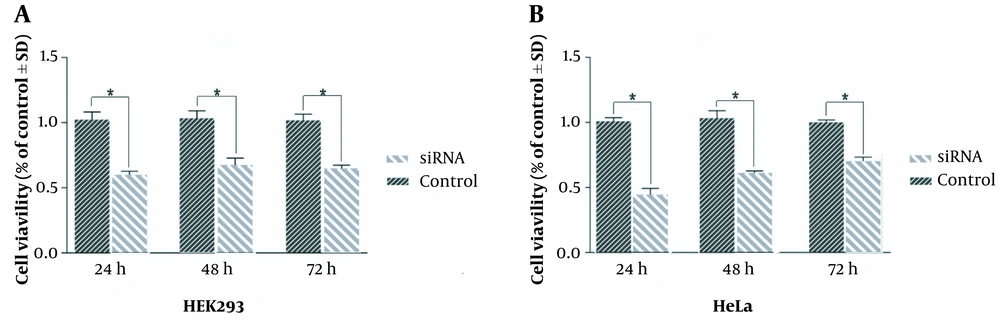
To elucidate the effect of HIF1α downregulation on cell viability, an MTT assay was conducted on HeLa and HEK293 cells. Statistical analysis revealed a notable reduction in the viability of HeLa cells by 59%, 66%, and 63% at 24, 48, and 72 hours after transfection, respectively (Figure 1A). Similarly, HEK293 cells exhibited a diminished viability of 48%, 61%, and 69% at corresponding time points (Figure 1B).
Statistical analysis indicated a significant decrease in HeLa cell viability by 59%, 66%, and 63% at 24, 48, and 72 hours after transfection, respectively (A); likewise, human embryonic kidney (HEK)293 cells demonstrated reduced viability of 48%, 61%, and 69% at the respective time intervals, as determined through MTT assay for cell viability (B). One-way analysis of variance (ANOVA) tests were performed on MTT results. Lines show the mean ± standard deviation (SD); *: P < 0.05.
3.2. Cell Cycle Perturbations Induced by HIF1α Knockdown
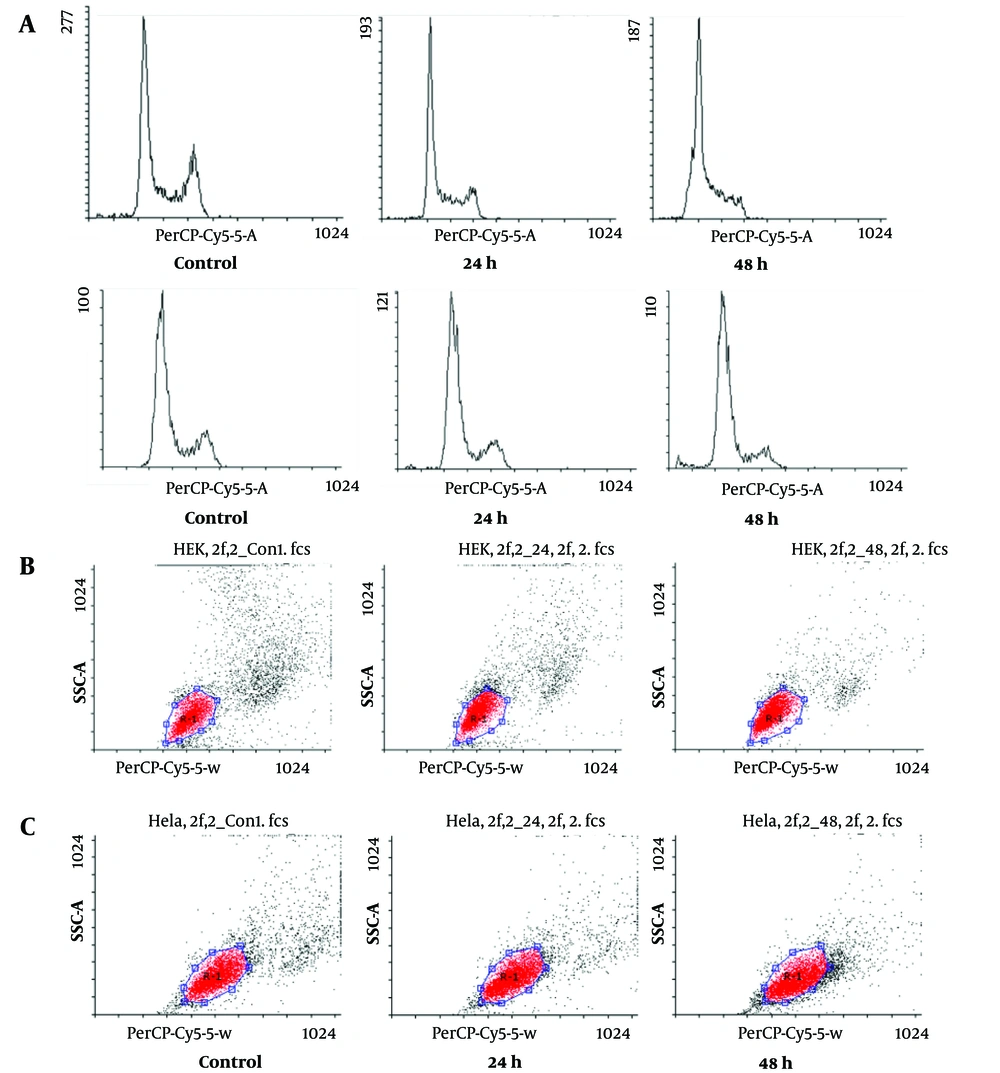
To assess alterations in the cell cycle induced by HIF1α knockdown, PI staining coupled with flow-cytometric analysis was employed on HeLa and HEK293 cells subjected to siRNA transfection. The results indicated an elevation in the percentages of cells in the sub-G1 phase and a concomitant reduction in the G2/M phase at 24 and 48 hours after transfection, compared to control cells (Figure 2A). The most substantial differences were observed at the 48-hour time point, where the percentages of HEK293 (Figure 2B) and HeLa (Figure 2C) cells in the sub-G1 phase increased (10- and 3-fold, respectively); nonetheless those in the G2/M phase decreased (10- and 3-fold, respectively) in comparison to control cells.
The cell cycle distribution was examined in human embryonic kidney (HEK)293 and HeLa cells transfected with si-hypoxia-inducible factor 1α (HIF1α) 24 and 48 hours after transfection. Cell cycle distribution was determined through propidium iodide (PI) staining and flow cytometric analysis; although there was a notable rise in cells in A, the sub-G1 phase, indicating apoptosis in both B, HEK293 and C, HeLa cells, there was a significant reduction in the percentage of cells in the G2/M phase for both HEK239 and HeLa cells, compared to control cells, at both 24 and 48 hours after transfection.
3.3. Impact of HIF1α Knockdown on XRCC4 and XRCC7 Expression
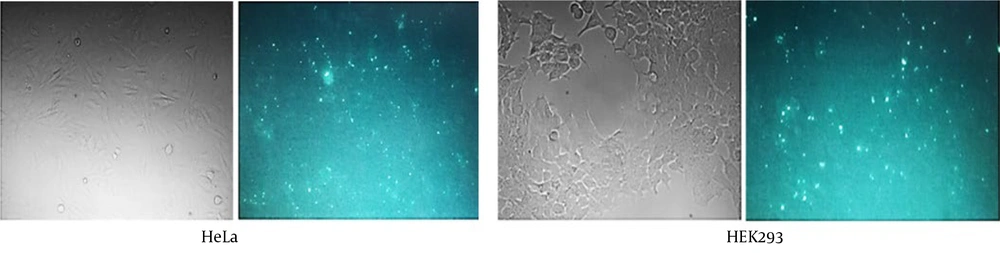
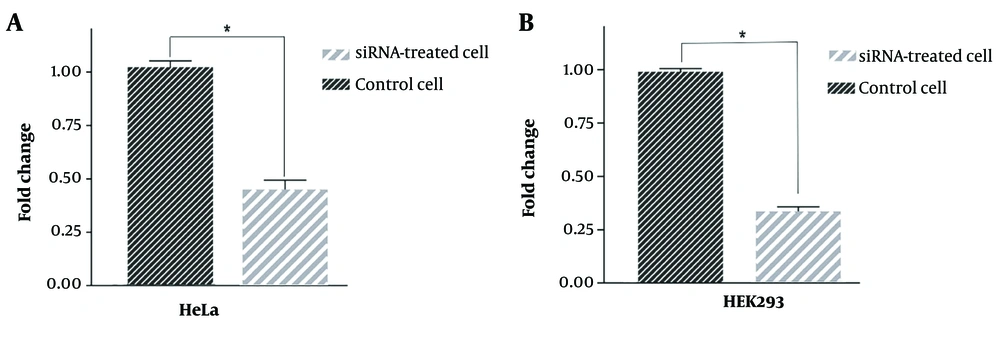
The efficiency of siRNA delivery, as observed through fluorescence microscopy (Figure 3), demonstrated successful transfection. The RT-qPCR technique was employed to quantitatively assess the effectiveness of the designed HIF1α-siRNA in downregulating its target mRNA. The results indicated a substantial reduction in HIF1α mRNA expression by 59% and 69% in HEK293 (Figure 4A) and HeLa (Figure 4B) cells, respectively.
Quantitative analysis of hypoxia-inducible factor 1α (HIF1α) messenger ribonucleic acid (mRNA) expression after downregulation using si-HIF-1α. Statistical analysis showed a significant decrease in the expression of HIF1α after 48 h both in human embryonic kidney (HEK)293 (A); and HeLa cells (B); *: P < 0.05.
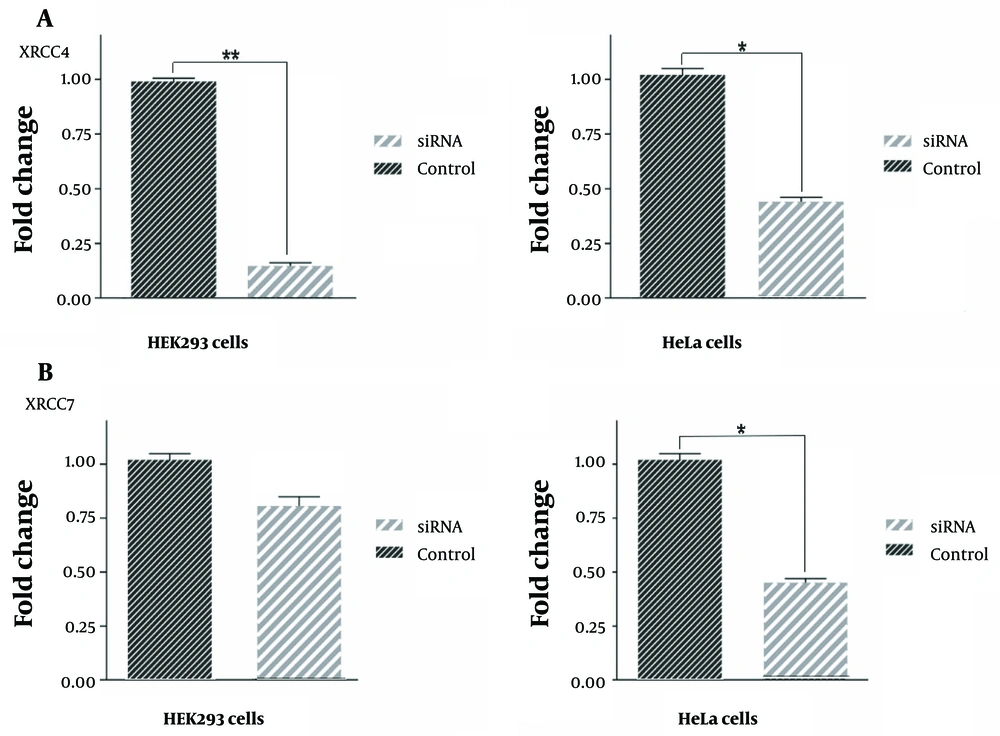
To investigate the influence of HIF1α downregulation, a pivotal transcription factor, on DNA repair genes, the expression levels of XRCC4 and XRCC7 were evaluated. The findings unveiled a significant decrease in XRCC4 mRNA expression by 86% and 57% in HEK293 and HeLa cells, respectively, 48 hours after transfection with HIF1α downregulation (Figure 5A). Conversely, XRCC7 exhibited no significant reduction in mRNA expression levels following HIF1α downregulation, observed in both HEK293 and HeLa cells (Figure 5B).
Inhibition of hypoxia-inducible factor 1α (HIF1α) leads to the downregulation of XRCC4 and XRCC7 genes. Quantitative analysis of XRCC4 and XRCC7 fold changes after downregulating of HIF1α gene using the small interfering ribonucleic acid (siRNA). Our results showed a significant decrease in the expression of the XRCC4 gene after transfection of both human embryonic kidney (HEK)293 and HeLa cell lines with si-HIF-1α (A); interestingly, these data did not replicate for the XRCC7 gene (B). Changes in gene expression compared to the control sample after the siRNA transfection in HEK293 cells had a slight decrease, which was not significant. However, these changes in HeLa cells showed a significant decrease; *: P < 0.05, **: P < 0.01.
3.4. In Silico Study
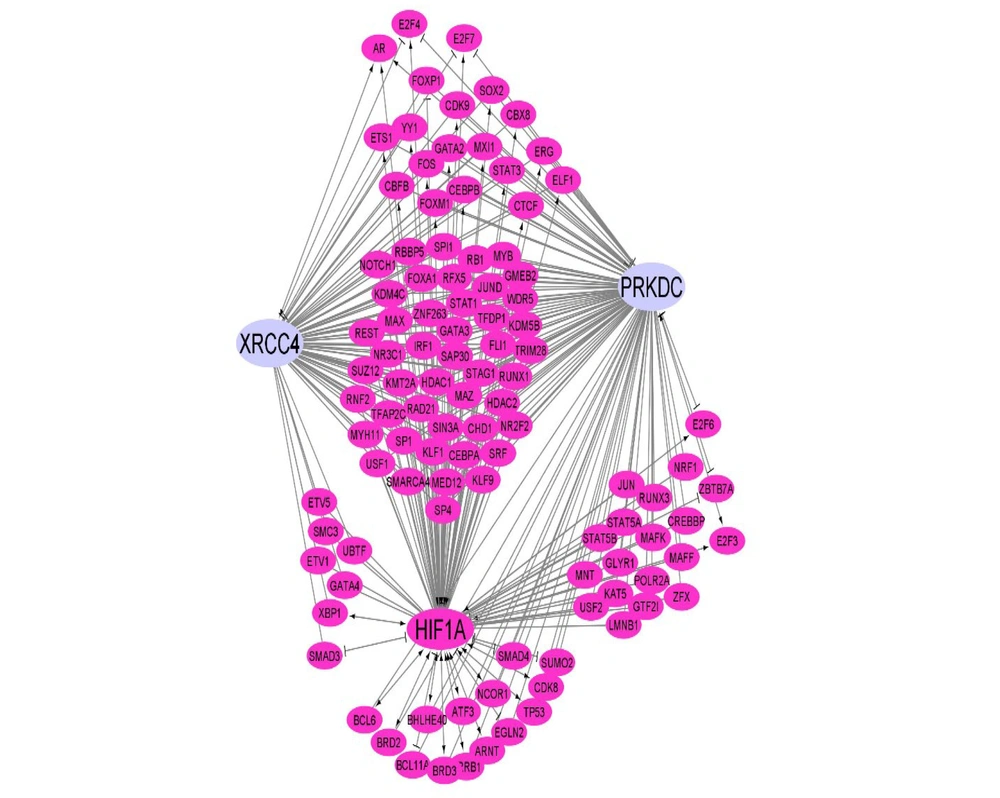
The examination of HIF1α binding sites within the promoter region (-5000 kb to 100 kb) of XRCC4 and XRCC7, utilizing the EPD, revealed 37 and 55 binding points (P = 0.01) for XRCC4 and XRCC7, respectively. In this investigation, a GRN or transcription factor-target network was constructed to depict the direct correlation of HIF1α with the XRCC4 and XRCC7 genes. Furthermore, endeavors were undertaken to illustrate the function of HIF1α as a hub gene, exerting influence on other transcription factors. This indirect influence portrays the examined transcription factor as a pivotal regulatory element capable of orchestrating gene regulation indirectly and serving as a regulatory hub within the network. The aforementioned findings not only emphasize the direct association of HIF1α with target genes but also elucidate its regulatory and indirect influencing role in the network (Figure 6).
This network represents the regulatory relationships governing the expression of XRCC4 and PRKDC (XRCC7) that are under the regulation of hypoxia-inducible factor 1 (HIF1)Α. The pink nodes are transcription factors (TFs), and the edges show the relationships. Arrow link: Activation; T shape link: Repression; and simple link: Unknown).
4. Discussion
Metastasis constitutes a complex series of events in cancer pathogenesis, leading to considerable complications for affected individuals (20). Notably, hypoxia emerges as a pivotal factor in heightening the metastatic potential of tumor cells, with concomitant evidence suggesting its role in inducing genomic instability (21, 22). Furthermore, investigations indicate alterations in the expression of DNA repair-related genes under hypoxic conditions (8, 23).
Glazer et al. posited that genomic instability arises from the modulation of DNA repair pathways, encompassing nucleotide excision repair, DNA mismatch repair, and homology-dependent repair (24). Notably, NHEJ predominates in DSB DNA repair, with numerous studies linking its inhibition to heightened carcinogenesis and genetic instability (25-27).
Serving as a master regulator in the hypoxic response, HIF1α orchestrates gene expression through its transcriptional activity. In a novel exploration, we assessed the impact of HIF1α downregulation on XRCC4 and XRCC7, pivotal genes in the NHEJ pathway. The functional experiments of the present study unveiled that siRNA-mediated HIF1α downregulation influences XRCC4 mRNA expression; however, XRCC7 remains unaffected. This finding suggests a role for XRCC4 in stabilizing and enhancing DNA ligase IV activity through physical interaction, which is crucial for the NHEJ repair system (28). Considering XRCC4's pivotal role in the late-stage NHEJ process, akin to the sealing of breaks, and its interaction with DNA ligase IV within the biological system, the use of XRCC4 knockout mice effectively replicates the observed deficiencies in DNA ligase IV knockout mice (29).
In the context of human tumor samples, Meng et al. reported diminished XRCC4 gene expression under hypoxic conditions, highlighting HIF1α's capacity to downregulate NHEJ pathway-related genes in both normal and cancer cells (8). A prior study by Chiappe-Gutierrez et al. established an increased XRCC1 expression in an experimental model of perinatal asphyxia (30).
Furthermore, Lo Nigro et al. demonstrated a significant upregulation of XRCC1, implicated in the repair of DNA single-strand breaks under hypoxic conditions (31). The diminished viability of HeLa and HEK293 cells following HIF1α down-regulation corroborated the findings by Mohebbi et al. (32) and Fernandes et al., (33) who reported reduced viability in glioma and retinoblastoma cells (32, 33). Consistent with Nakamura et al., a notable increase in sub-G1 cells, particularly 48 hours after transfection, suggested apoptosis in both HeLa and HEK293 cells (34). Intriguingly, a decrease in G2/M arrest at 48 hours after transfection, likely linked to XRCC4 inhibition through HIF1α downregulation, implies a potential correlation between XRCC4 inhibition in the NHEJ repair pathway and apoptosis induction.
Furthermore, cell cycle assessments demonstrated that HIF1α knockdown led to increased apoptosis, aligning with the findings of GD He et al., who emphasized the promotion of apoptosis in pancreatic cancerous BxPC-3 cells with HIF-1α knockdown (35). Although some studies have posited hypoxia-induced cell death through p53 stabilization by HIF1α (36), the results of the present study propose an alternative mechanism involving the downregulation of XRCC4, a pivotal player in the NHEJ pathway. Despite the bioinformatics analyses of the current study revealing 37 and 55 HIF1α-binding sites in the promoters of XRCC4 and XRCC7 genes, respectively, it is noteworthy that the experimental reduction of HIF1α expression did not yield a discernible impact on XRCC7 expression levels. This observation implies that XRCC7 might not be solely under the regulation of HIF1α in this pathway, underscoring the need for additional investigations into the regulatory networks influencing these observed changes.
The intricate interplay of transcription factors and their binding to gene transcription control regions complicates the regulation of gene expression. Consequently, in this network, transcriptional co-regulators emerge as equally important modulators of the responses observed in this study. However, establishing a universal molecular connection between HIF1α, XRCC4, and XRCC7 proves challenging. Therefore, further studies are imperative for unraveling this complex transcription factor-regulator network.
In conclusion, transcription factors serve as pivotal mediators in effective intervention against cancer diseases, orchestrating cell fate decisions. The results of the current study unveil that the knockdown of HIF1α, a master regulator, influences the NHEJ-related XRCC4 gene, diminishes cell viability, and enhances the apoptotic potential of HeLa and HEK293 cells. These findings suggest that HIF1α could serve as a promising target for cancer therapy.