1. Background
Maintaining the nucleotide pool, ensuring genomic DNA integrity, and having robust repair systems are crucial for life (1). Loss of DNA integrity and chemical modifications of nucleotides can lead to various human diseases, including neurological disorders (2). Such modifications are a primary pathway for the accumulation of abnormal DNA bases, often caused by endogenous factors like reactive oxygen species (ROS) (3). To counter these harmful effects, evolution has developed an enzyme called inosine triphosphatase (ITPA, EC3.6.1.19) (4), which plays a significant role in the immune system (5).
The human ITPA gene contains two functional SNPs: A missense variation in exon 2, 94C->A (rs1127354/dominant wild-type allele), which results in a P32T substitution and complete ITPAse deficiency, leading to abnormal ITP accumulation in erythrocytes (6). This variation affects the protein in three ways: It reduces the expression of the ITPA transcript, decreases enzymatic activity, and lowers protein stability (4). The other SNP is a splicing variant in intron 2, IVS2 + 21A → C (rs7270101/dominant mutant allele), which independently decreases gene expression, causing inosine deficiency (7). Homozygosity for the ITPA IVS2+21C allele results in a 40% reduction in ITPAse activity in red blood cells. Additionally, compound heterozygotes (ITPA 94A/IVS2 + 21C) exhibit only 10% of normal activity (4). This SNP is also linked to potentially adverse drug reactions to azathioprine (8), an immunosuppressive drug used in autoimmune diseases, such as multiple sclerosis (MS) (9).
ITPA has two transcript variants. Transcript variant 1 is the longest transcript (isoform a), encoding the longest isoform, while transcript variant 2 uses an alternate in-frame splice site in the 5' coding region compared to variant 1, resulting in a shorter protein (isoform b) than isoform a (6).
The rs1127354 and rs7270101 SNPs in the ITPA gene have been linked to varying levels of pathogenicity potential and allele distribution in immune disorders across different populations (10, 11). The rs1127354 SNP, located in the ITPA gene, has been associated with altered enzyme activity, which may impact purine metabolism and immune responses (10, 12). Research indicates that certain genotypes of rs1127354 could influence susceptibility to autoimmune diseases, including MS (12). The prevalence of these variants differs among populations, with some ethnic groups exhibiting higher or lower frequencies of specific genotypes (13). Understanding the pathogenic potential and frequency distribution of these variants in diverse populations is crucial for elucidating their role in disease susceptibility and for developing personalized medicine approaches for MS and other autoimmune disorders.
For instance, these SNPs are associated with the metabolism of ribavirin (RBV) and can affect the efficacy of antiviral therapies, particularly in chronic hepatitis C virus (HCV) infections (13). They also contribute to adverse effects, such as hemolytic anemia, in HCV patients undergoing RBV-based antiviral therapy (13). Additionally, the rs1127354 SNP has been linked to neutropenia in both general populations and children, indicating a broader impact on immune-related issues (14). The prevalence of these SNPs varies among different populations with immune diseases. For example, the rs1127354 CC genotype is found in approximately 72.8% of Japanese and 81.0% of Korean individuals, highlighting its relatively high prevalence in these regions (13). Similarly, in Northeast China, the rs1127354 CC genotype is present in 76.2% of individuals with HCV genotype 1 and 77.6% of those with non-genotype 1 HCV, which is comparable to the frequencies observed in Japanese and Korean populations (13).
2. Objectives
The aim of this investigation was to examine the association between two specific genetic variations (rs1127354 and rs7270101 SNPs) and the expression of the ITPA gene in patients with MS. The ITPA gene is critical for maintaining cellular homeostasis by regulating intracellular levels of inosine triphosphate (ITP) and diphosphate (IDP). Dysregulation of ITPA can lead to the accumulation of ITP and IDP (15), potentially disrupting immune cell functions (16). This study provides a novel perspective by evaluating the connection between rs1127354 and rs7270101 SNP genotyping and ITPA gene expression in individuals with MS. Although previous research has investigated genetic factors associated with MS susceptibility, the specific relationship between these SNPs and ITPA gene expression in MS has not been extensively explored. By focusing on these particular SNPs and their potential impact on ITPA regulation, this research aims to deepen the understanding of the molecular mechanisms underlying MS etiology, potentially paving the way for the development of targeted therapies and personalized treatments for affected individuals.
3. Methods
3.1. Study Subjects
This study included 112 patients with MS and 109 healthy controls who were genotyped for the rs1127354 and rs7270101 SNPs. From these, 32 patients and 30 controls were randomly selected for analysis of the relative changes in the expression of two ITPA transcript variants. Multiple sclerosis patients were recruited from Sina Hospital and diagnosed by a specialist physician based on the McDonald criteria (17) and MRI findings. Control subjects, matched for sex and age, were volunteers without neurological or other diseases such as cancer or diabetes. Informed consent was obtained from all participants, and the study was approved by the Ethics Committee of Tarbiat Modares University under the ethical approval code IR.TMU.REC.1395.373. The sample size was calculated based on the study by Sahraian et al. (18).
3.2. Genotyping
Blood samples were collected in 5 ml EDTA tubes, and genomic DNA was extracted using the DNG PlusTM Kit (Cinnagen, Iran). Genotyping for rs1127354 and rs7270101 was performed using the Mismatch PCR-RFLP (PCR-Restriction Fragment Length Polymorphism) method. The primers used were:
- For rs1127354: Forward: 5′-CAGGTCGTTCAGATTCTAGGAGAAAAGT-3′, reverse: 5′-CAAGAAGAGCAAGTGTGGGACAAG-3′
- For rs7270101: Forward: 5′-AAATTGACCGTATGTCTCTGGAATGTTT-3′, reverse: 5′-CAAGAAGAGCAAGTGTGGGACAAG-3′
The mismatched nucleotides in the forward primers are underlined. The PCR protocol included denaturation at 94°C for 5 minutes, followed by 35 cycles of 94°C for 30 seconds, annealing at 53°C for 30 seconds, 72°C for 30 seconds, and a final extension at 72°C for 5 minutes. The PCR products were digested with XmnI restriction enzyme (Takara Bio Inc., Japan) at 37°C and analyzed by electrophoresis on 12% PAGE (polyacrylamide gel electrophoresis). The amplicon sizes for rs1127354 and rs7270101 were 256 bp and 204 bp, respectively. Selected specimens from each genotype were validated by sequencing with an ABI automated DNA sequencing system (Macrogen, Korea).
3.3. ITPA Expression
Blood samples (5 mL) were collected from 32 MS patients and 30 controls. Peripheral blood mononuclear cells (PBMCs) were isolated using density gradient centrifugation with Ficoll/Paque solution (Lympholyte, Netherlands). Total RNA was extracted from PBMCs using the RNXTM-plus reagent (Cinnagen, Iran) according to the manufacturer's instructions and treated with DNase I (Sigma) at 37°C for 30 minutes. RNA concentration, quality, and integrity were verified using spectrophotometry and agarose gel electrophoresis. cDNA synthesis was performed using RevertAidTM Reverse Transcriptase (Fermentas, Canada), with 3 µg of total RNA, random hexamer primers (MWG, Germany), and oligo (dT) primers.
The relative expression of ITPA transcript variant 1 (forward: 5΄-AAGAAGCTGGAGGAGGTCG-3΄ and reverse: 5΄-TCCAAGGGCATTGAAGCACAG-3΄) and variant 2 (forward: 5΄-GGCGGCCTCATTGGTCGTTC-3΄ and reverse: 5΄-TCCAAGGGCATTGAAGCACAG-3΄) was measured using an Applied Biosystems 7500 real-time PCR system (Applied Biosystems/MDS SCIEX, USA). The reactions included 10 µL of SYBR Green I master mix (Takara, Japan), 200 nM of each primer, and 10 ng of cDNA. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (reverse: 5΄-GAGTCCTTCCACGATACC-3΄ and forward: 5΄-CCATGAGAAGTATGACAAC-3΄) was used as the reference gene.
3.4. Statistical Analysis
The results were analyzed using the SPSS software package (version 20.0). The chi-square test was used to evaluate Hardy-Weinberg equilibrium (HWE), compare allele-genotype frequencies, and assess genotype-phenotype associations. A significance level of 0.125 was selected with Bonferroni's correction to achieve the traditional P-value of ≤ 0.05. Additionally, the odds ratio (OR) and 95% confidence interval (CI) were calculated for both the patient group and healthy controls. Haplotype frequencies were estimated using PHASE (version 2.1.1) software.
The independent t-test and ΔCt methods (19) were used to compare the expression levels of ITPA variants between controls and patients. The correlation between genotypes and gene expression levels was evaluated using a two-sided Mann–Whitney U test with GraphPad Prism 5. Pearson's correlation coefficient was used to analyze the correlation between normalized expression of ITPA transcript variants and participants’ age and disease duration.
4. Results
4.1. Participants’ Characteristics
Table 1 presents the characteristics of the patients and controls.
Abbreviations: RR, relapsing remitting; SP, secondary progressive; PP, primary progressive; EDSS, expanded disability status scale.
a Values are expressed as mean ± SD.
4.2. ITPA Polymorphisms and the Risk of Multiple Sclerosis
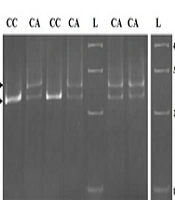
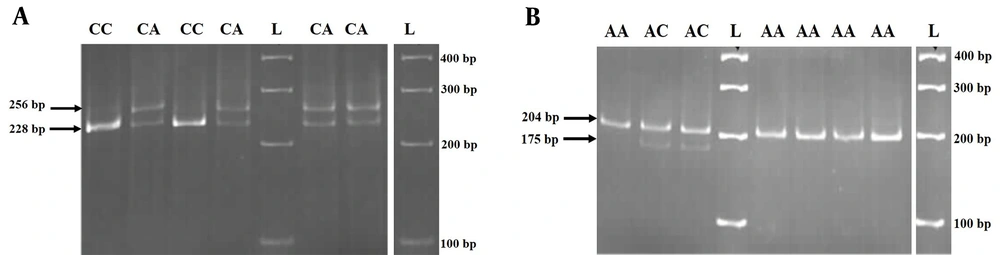
Two SNPs in the ITPA gene were genotyped in both the patient and control groups using mismatched PCR-RFLP methods. Different genotypes for rs1127354 and rs7270101 were observed on a 12% acrylamide gel (Figure 1A and B). Sequencing results confirmed the genotyping. All genotyped SNPs were in Hardy-Weinberg equilibrium (HWE) in both groups. We compared the allele frequencies and genotype distributions for rs1127354 and rs7270101 between patients and controls (Table 2). No significant differences were found between patients and controls at the allele or genotype levels. The frequency of the minor rs1127354 allele (A) was 9% in the patient group and 7% in the control group (P = 0.53). The allelic frequency for rs7270101 (A, C) was 92% and 8% in the patient group vs. 94% and 6% in the control group, respectively (P = 0.41). No significant associations were found between these two polymorphisms and MS risk in either the female or male groups.
The results of mismatch PCR-RFLP by the XmnI enzyme on a 12% polyacrylamide gel. A, the genotyping of rs1127354. There was no restriction site at the C/A locus. The T nucleotide near the SNP was replaced with an A nucleotide in the PCR reaction using a mismatched forward primer; therefore, the XmnI enzyme could cut wild-type (CC) or heterozygote (CA) genotypes, and 228 bp and 28 bp fragments were produced; B, the genotyping of rs7270101. There was no restriction site at the A/C locus. The TTT nucleotides near the SNP were replaced with GAA nucleotides in the PCR reaction by using a mismatched forward primer; therefore, the XmnI enzyme could cut mutant (CC) or heterozygote (AC) genotypes, and 176 bp and 28 bp fragments were produced. L: Ladder (molecular marker).
| SNP | Genotype and Allele | Patient (n = 112) | Control (n = 109) | Odds Ratio | Confidence Interval | ꭓ2 | P-Value |
|---|---|---|---|---|---|---|---|
| rs1127354 | CC | 93 (83) | 94 (86) | 0.77 | (0.36 - 1.67) | 0.41 | 0.52 |
| CA | 18 (16) | 14 (13) | 1.28 | (0.58 - 0.28) | 0.40 | 0.52 | |
| AA | 1 (1) | 1 (1) | 1.16 | (0.07 - 18.82) | 0.11 | 0.91 | |
| Maj. Allele: C | C | 91 | 93 | - | - | - | - |
| Min. Allele: A | A | 9 | 7 | 0.79 | (0.39 - 1.62) | 0.39 | 0.53 |
| rs7270101 | AA | 95 (85) | 97 (89) | 0.70 | (0.31 - 1.59) | 0.70 | 0.40 |
| AC | 17 (15) | 12 (11) | 1.41 | (0.62 - 3.18) | 0.70 | 0.40 | |
| CC | 0 (24) | 0 (0) | - | - | - | - | |
| Maj. Allele: A | A | 92 | 94 | - | - | - | - |
| Min. Allele: C | C | 8 | 06 | 0.72 | (0.33 - 1.58) | 0.65 | 0.41 |
a Values are expressed as No. (%) or %.
b The chi-square test was employed to assess if there were any significant variances (P-values) between the patient group and the control group. The genotype distribution of SNPs did not show any significant differences between MS patients and the control group.
4.3. Haplotype Analysis and Risk of Multiple Sclerosis
We also assessed the haplotypes of the two genotyped SNPs concerning MS susceptibility. Four possible haplotypes were identified in our dataset, with one being very uncommon. No significant correlation was found between MS and any of the haplotypes (P = 0.62, Table 3).
| Variables | rs1127354 C/A | rs7270101 A/C | Frequency of Controls | Frequency of Patients | OR | P-Value |
|---|---|---|---|---|---|---|
| Hap 1 | C | A | 0.85 | 0.84 | 1 | - |
| Hap 2 | C | C | 0.047 | 0.042 | 0.85 | 0.67 |
| Hap 3 | A | A | 0.096 | 0.11 | 0.91 | 0.75 |
| Hap 4 | A | C | 0.0004 | 0.0000 | 0 | - |
a The chi-square test was employed to assess if there were any significant differences (P-value) between the group of MS patients and the group of healthy control individuals.
b There is no significant relationship between any of the haplotypes and MS (P-value: 0.05).
4.4. ITPA Expression
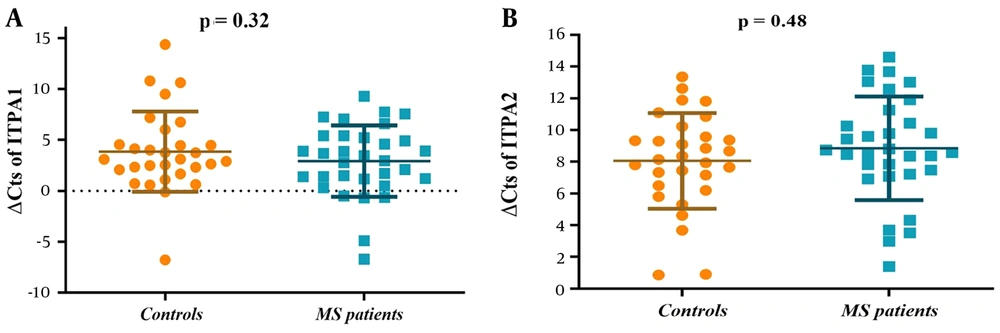
The expression analysis of two transcript variants of ITPA was measured as ΔCt. qRT-PCR analysis showed no significant difference in the expression of ITPA variant 1 and variant 2 between MS patients and controls (P = 0.32 and P = 0.48, respectively) (Figure 2A and B).
The expression analysis of the ITPA gene. Normalized expression (∆Cts) of the ITPA transcript variants 1 (A) and 2 (B) showed no difference in the PBMCs of MS patients compared to controls. The expression levels of transcripts in each sample were normalized to GAPDH expression. (The normalized expression is ∆Ct = Ct Target gene – Ct Housekeeping gene). The t-test was used to examine the difference in gene expression between the two groups.
4.5. The Effect of the rs1127354 SNP on ITPA Expression
We grouped heterozygous and homozygous cases for the ITPA A risk allele (AA and CA) and compared them with homozygous individuals for the protective C allele. Our data indicated that the rs1127354 A allele in the ITPA gene did not affect the expression of ITPA mRNA in MS patients (P = 0.61) or control subjects (P = 0.45).
4.6. Correlation of ITPA Transcript Variants' Expression Levels with Demographic and Clinical Characteristics
Our analyses showed no correlation between the normalized expression of ITPA transcript variant 1 and EDSS (Expanded Disability Status Scale) scores, disease duration, or patient age. The EDSS is an ordinal scale providing a standard clinical assessment to evaluate the level of disability in MS individuals (Validity of Outcome Measures, Clinical Review Report, NCBI). Similarly, no correlation was observed for the expression levels of ITPA transcript variant 2. However, the levels of ITPA transcript variant 2 positively correlated with disease duration in patients (Table 4).
| Correlations | Age | Disease Duration | EDSS | |||
|---|---|---|---|---|---|---|
| r | P-Value | r | P-Value | r | P-Value | |
| ITPA transcript variant 1 | -0.004 | 0.983 | 0.093 | 0.613 | -0.046 | 0.015 |
| ITPA transcript variant 2 | 0.027 | 0.882 | 0.375 | 0.035 a | 0.313 | 0.081 |
a Correlation is significant at P < 0.05.
Furthermore, we studied the association between the expression levels of the evaluated genes. A significant positive association was observed between the expression levels of ITPA transcript variants 1 and 2 in MS patients (r = 0.426, P = 0.015).
5. Discussion
In this study, we focused on genotyping two SNPs in the ITPA gene and examining the expression of two ITPA mRNA variants. The selection of rs1127354 and rs7270101 SNPs for this investigation was based on pioneering research suggesting their potential relevance to MS and the ITPA gene. These specific SNPs have been implicated in several immune system disorders and have shown correlations with altered gene expression and other conditional probabilities (12). Thus, the choice of rs1127354 and rs7270101 SNPs for genotyping in MS patients was driven by their hypothesized practical significance and potential role in regulating ITPA expression and disease etiology.
Our results showed no significant association between the two SNPs and MS, nor between the two SNPs and gene expression. Previous studies have investigated the relationship between ITPA polymorphisms and various diseases, including MS (20), hemolytic anemia (21), acute lymphoblastic leukemia (22), inflammatory bowel disease (23, 24), and epileptic encephalopathy (25). However, only one study has suggested that ITPA may play a potential role in immunity. The two proteins encoded by ITPA were initially identified by cytotoxic T-cells. Although alternative splicing is one mechanism that produces these immune proteins, this hypothesis is not yet fully understood (5).
Furthermore, it is hypothesized that ITPA is crucial for maintaining the housekeeping functions of immune cells, including eosinophils and macrophages. Nevertheless, little is known about ITPA activity in these cells. Supporting this hypothesis, in two other inherited disorders—purine nucleoside phosphorylase and adenosine deaminase deficiency—ITPA deficiency results in severe immune diseases, characterized by severe T-cell immunodeficiency and combined immune deficiency (5). Additionally, SNPs in ITPA, including the P32T variant, have been observed as potential susceptibility factors for tuberculosis, possibly leading to immune system deficiencies. It is crucial to note that some alleles may influence the regulation (transcription and/or translation) of ITPA, decreasing its expression. Reduced expression diminishes the host immune system's response against tuberculosis infection in young patients (26).
Moreover, in silico studies using the UCSC genome browser (http://genome.ucsc.edu/) have shown that ITPA expression is high in immune cells (26). Furthermore, the knockdown of ITPA induces apoptosis in SKBR3 cancer cells (27). Although the role of the Inosine Triphosphatase (ITPA) gene product in humans is not yet fully understood, it is essential for genome stability. In fact, ITPA is involved in the DNA repair system, preventing the accumulation of deaminated nucleotides in DNA and RNA (28), and functions as an oxidative nucleotide scavenger (20).
As Kumar et al. stated, the recycling of purines trapped in the form of ITP and the protection of the cell from the accumulation of non-canonical nucleotides, such as ITP, dITP, or XTP, which may be incorporated into DNA and RNA, is the putative role of ITPAse. These rogue nucleotides can increase the mutational load in genetic material, gradually leading to the loss of genome integrity, with various outcomes (29). Additionally, recent studies have indicated that oxidative stress is involved in the pathophysiology of MS progression and lesion generation (30). Predominantly, ROS are known to mediate immune cell trans-endothelial migration and induce dysfunction in the blood-brain barrier, particularly during disease initiation and lesion formation (30). Furthermore, previous studies have explored the relationship between ITPA polymorphisms and the side effects of drugs on MS. For instance, azathioprine is widely used as an anti-inflammatory and immunosuppressive drug. ITPAse metabolizes this agent after it enters the body (4). Reduced ITPAse function leads to the accumulation of ITP in erythrocytes (31). Therefore, under conditions of cellular stress, a deficiency in this enzyme can be problematic (32).
Generally, the rs1127354 and rs7270101 SNPs in the ITPA gene exhibit pathogenic features in immune disorders, particularly in adverse effects related to antiviral therapeutics such as RBV (12). The frequency of these SNPs varies among different populations (13, 14), highlighting the importance of understanding genetic variations in immune-linked conditions for targeted and personalized treatment approaches.
Although our study did not find an association between the expression of ITPA and MS, it is possible that ITPA expression in immune cells is influenced by rs1127354 and rs7270101 SNPs. Given the association of SNPs with gene pools in different populations, further research with larger sample sizes and diverse ethnic and racial populations is necessary. Additionally, future studies should explore biologically significant interactions between genes that confer susceptibility to MS, as well as the levels and activities of the ITPA protein (4).
5.1. Conclusions
This investigation advances the field of MS research by demonstrating no link between rs1127354 and rs7270101 SNP genotyping and ITPA expression in MS patients. These findings enhance our understanding of the molecular mechanisms underlying MS pathogenesis by indicating that these SNPs may not play a role in regulating ITPA expression or in the susceptibility and progression of MS. This work contributes to a deeper understanding of the genetic factors influencing MS and underscores the importance of targeted therapies and personalized medicine approaches in treating this complex autoimmune disease. Ultimately, this research improves our understanding of the complex interplay between genetic factors and MS, potentially opening new avenues for precision medicine approaches in MS management.