1. Background
Lung adenocarcinoma (LUAD) accounts for the majority of non-small-cell lung cancer (NSCLC), one of the leading causes of cancer morbidity worldwide. As with other cancers, the exact molecular mechanisms of LUAD have not been fully understood (1). Recently, long non-coding RNAs (lncRNAs) have emerged as important targets in cancer research. Long non-coding RNAs are a class of regulatory RNAs that can function as oncogenes or tumor suppressors and serve as diagnostic or prognostic cancer biomarkers (2).
Long non-coding RNAs regulate the proliferation, invasion, and chemoresistance of cancer cells, primarily through their involvement in gene expression regulation. Gene expression regulation is a complex process with multiple control levels, and lncRNAs play a role in influencing it (3). Long non-coding RNAs can modify chromatin by interacting with chromatin modifiers or directly binding to DNA. Additionally, they can interact with transcription factors (TFs) and RNA polymerases, affecting their functions. In the cytoplasm, lncRNAs exert post-transcriptional, translational, and post-translational regulation on gene expression. Their regulatory mechanisms include interacting with mRNAs, sponging microRNAs, interacting with the translation machinery, and binding to peptides (4, 5).
As a result, the dysregulation of lncRNAs can lead to disruptions in the expression levels of various genes. Gene expression dysregulation is a common molecular feature of cancers and can be utilized for cancer diagnosis and treatment (6).
Long non-coding RNAs can have both oncogenic and tumor-suppressive effects in cancer (7). LINC00525 is an upregulated and oncogenic lncRNA in LUAD and other cancers, such as colorectal cancer and spinal chordoma (8-10). LINC00525 is associated with high rates of cell proliferation and metastasis in these cancers (9). Several molecular mechanisms have been proposed for LINC00525, including reducing the transcription and stability of p21 (a cell cycle blocker) in LUAD and sponging miR-31-5p and miR-125a-5p in spinal chordoma (9, 10). In colorectal cancer, overexpression of LINC00525 leads to apoptosis resistance and cell cycle progression through the suppression of tumor suppressor genes P15, P21, P27, and KLF2 (8). Additionally, LINC00525 overexpression promotes metastasis, stemness, and chemoresistance in cancer cells (11, 12). Therefore, understanding the downstream regulatory networks of LINC00525 is crucial, and targeting LINC00525 could be a promising strategy in cancer therapy.
2. Objectives
This study utilized in silico data to identify genes whose expression was altered following LINC00525 knockdown in a LUAD cell line. The gene expression profile GSE171460 was used to determine the differentially expressed genes (DEGs) after silencing LINC00525 in LUAD A549 cells. The aim was to identify genes related to LINC00525 in LUAD cells and explore potential molecular mechanisms that could be targeted in therapeutic approaches. Additionally, the study investigated related signaling pathways and their effects on cellular processes. The correlation between LINC00525 and the expression of hub genes, as well as the regulatory network of related TFs, was also evaluated.
3. Methods
3.1. Datasets and Data Processing
Using the UALCAN database, the expression of LINC00525 was analyzed across various cancer types, including LUAD. RNA-seq LUAD data were obtained from TCGA using R software, comprising 541 cases and 59 matching adjacent normal tissues. The ROC curve and expression level of LINC00525 were examined using GraphPad Prism 9. The GSE171460 data package, containing microarray data from human LUAD A549 cells, was downloaded from the GEO database. In this dataset, LINC00525 was downregulated using siRNA in LUAD cells, while scramble siRNA was transfected into A549 cells as a negative control. Differentially expressed genes between the control and LINC00525 knockdown groups were identified using GEO2R online tools and the R program.
3.2. Pathway Enrichment Analysis
The R package enrichR was used for Reactome_2022 analysis of DEGs in A549 cells. The plotEnrich package was employed to visualize the results, with an adjusted P-value of ≤ 0.05.
3.3. Hub Genes Identification
The STRING database was used to construct a protein-protein interaction (PPI) network of DEGs, with interactions having scores > 0.4 considered statistically significant. The results were visualized using CytoScape (version 3.6.1) software. To identify the hub genes, CytoHubba, a plug-in of CytoScape, was employed, utilizing the degree algorithm for selection. The correlation between the expression of hub genes and LINC00525 was evaluated using the GEPIA database.
3.4. Prediction of Transcription Factors Which Can Be Interacted with LINC00525
The RNAInter database was used to identify TFs that interact with LINC00525. The TRRUST database was employed to predict target genes regulated by the identified TFs (adjusted P-value ≤ 0.05). The overlap between the predicted target genes and the differentially expressed mRNAs in A549 cells was analyzed using a Venn diagram.
4. Results
4.1. LINC00525 Was Considerably Upregulated in Lung Adenocarcinoma
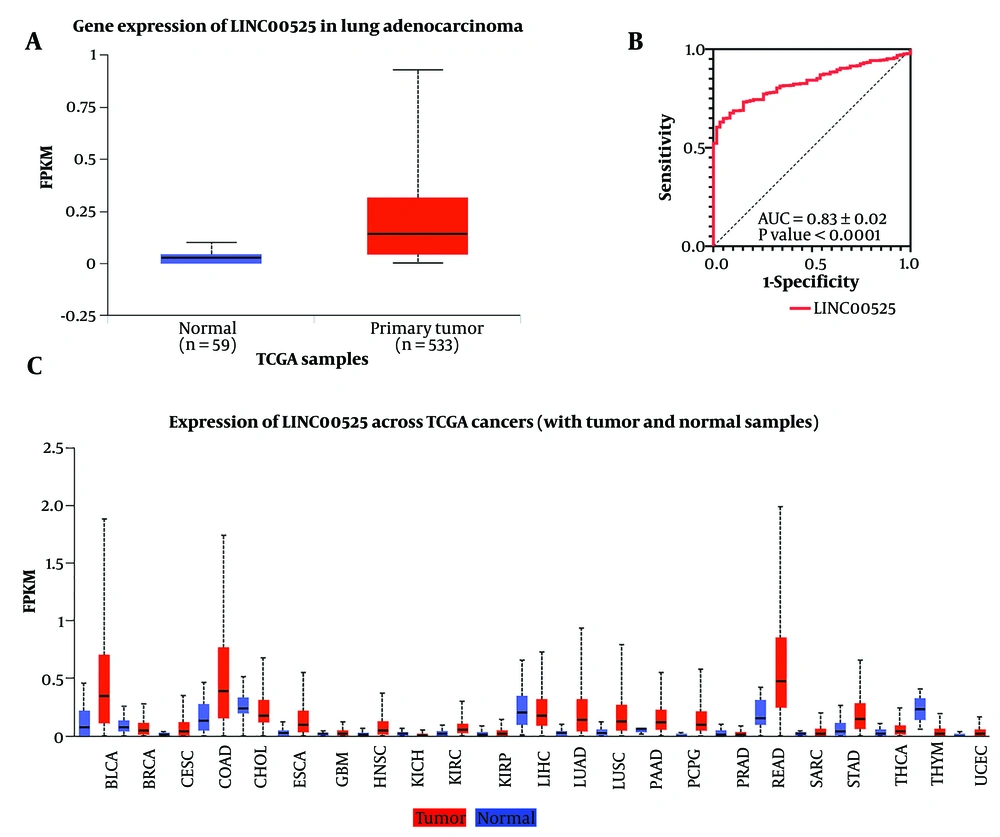
Analysis of RNA-seq datasets from the TCGA revealed that LINC00525 was expressed at significantly higher levels in tumors compared to normal tissues (Figure 1A). ROC curve analysis further confirmed the potential of this lncRNA to serve as a diagnostic biomarker in LUAD (Figure 1B). According to a pan-cancer investigation using the UALCAN database, LINC00525 expression was upregulated in various cancer types, particularly in LUAD (Figure 1C).
Expression pattern and diagnostic value of LINC00525. A, significant expression difference (P-value cutoff = 0.05) of LINC00525 in tumor compared to normal tissue according to UALCAN; B, the ROC curve of LINC00525 in LUAD. AUC was 0.91 with 95% CI. Red represents the sensitivity curve, and black indicates the identification line. The X axis shows the false positive rate, presented as “1-Specificity”. The Y axis indicates the true positive rate, shown as “Sensitivity”; C, overexpression of LINC00525 was shown in several cancers, most notably LUAD tissue according to UALCAN.
4.2. Changes of mRNA Profile After the Silencing of LINC00525
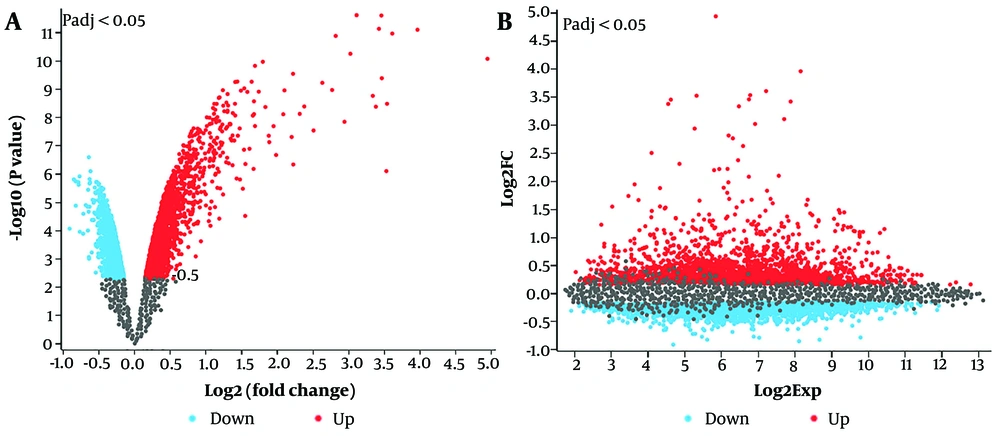
A total of 3,093 differentially expressed mRNAs (|log2FC| ≥ 0.1 and adjusted P-value ≤ 0.05) were identified in the GSE171460 dataset. Among these, 1,360 DEGs were upregulated, and 1,733 were downregulated in the lncRNA downregulated cells compared to control LUAD cells (Figure 2A and B). The MX1 gene, with a logFC = 4.94, was the most upregulated, while the LOC102724250 gene, with a logFC = -0.9, was the most downregulated. After integrating the DEGs, genes with |log2FC| ≥ 0.5 were selected for further analysis.
4.3. Pathway Enrichment Analysis, Protein-Protein Interaction Network Construction of Differentially Expressed Genes, and Selection of Hub Genes
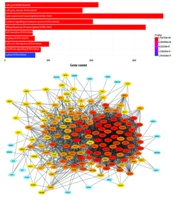
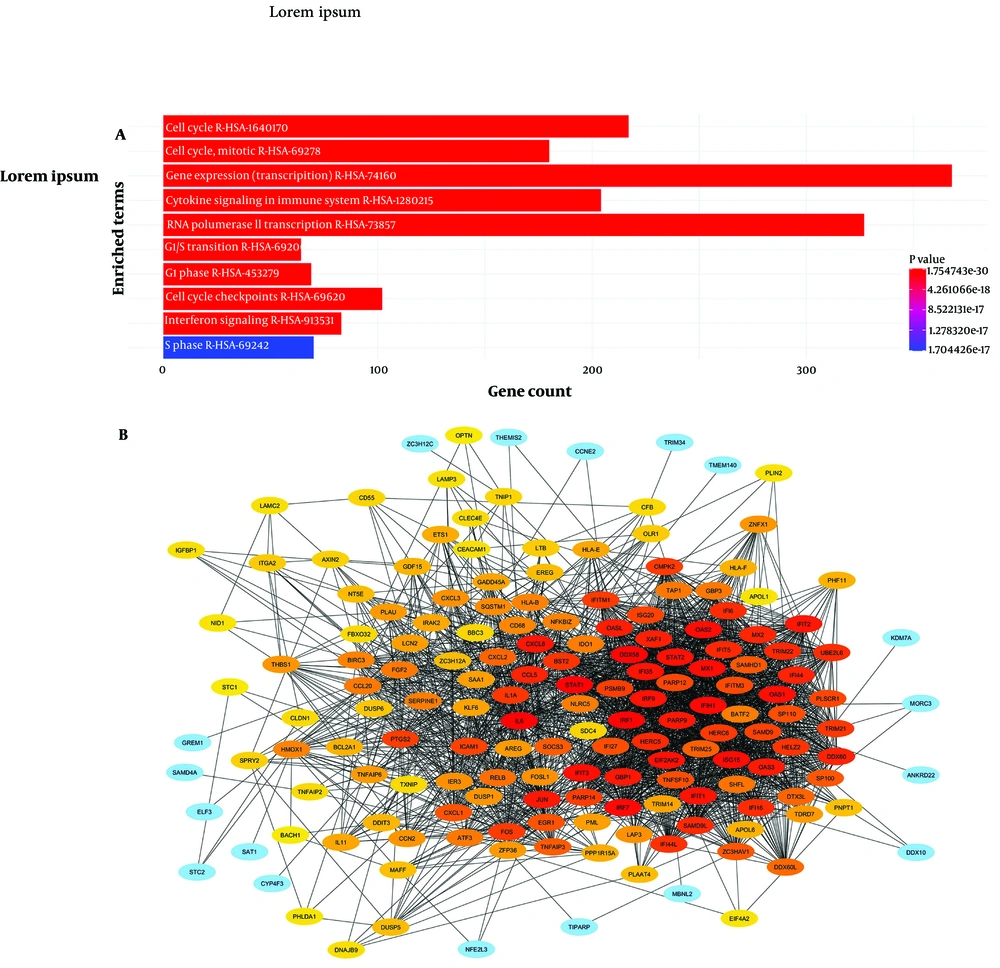
In the Reactome_2022 analysis, the most significantly enriched pathway for DEGs was the cell cycle (Figure 3). In addition to the cell cycle, cytokine signaling was also among the significantly enriched pathways following LINC00525 silencing. The protein interaction network of the DEGs was constructed using STRING. Based on the Degree method of CytoHubba in Cytoscape, 11 hub genes were identified from the STRING data (Table 1).
Pathway enrichment analysis, protein-protein interaction (PPI) network construction of differentially expressed genes (DEGs). A, significant pathways related to DEGs; and B, PPI network for the DEGs of A549 cells (downregulated LINC00525 compared to control) in Cytoscape, genes with more interactions are depicted with the high intensity of red color.
| Rank | Gene Name | Score | logFC (in LINC00525 Downregulated Compared to Control) |
|---|---|---|---|
| 1 | STAT1 (signal transducer and activator of transcription 1) | 92 | 1.31 |
| 2 | IL6 (interleukin 6) | 90 | 1.34 |
| 3 | IRF1 (interferon regulatory factor 1) | 84 | 0.7 |
| 4 | DDX58 (DEAD (Asp-Glu-Ala-Asp) box polypeptide 58) | 80 | 2.82 |
| 5 | IRF7 (interferon regulatory factor 7) | 77 | 2.08 |
| 6 | IFIH1 (interferon induced with helicase C domain1) | 74 | 0.5 |
| 7 | MX1 (MX dynamin like GTPase 1) | 73 | 4.94 |
| 8 | OAS2 (2'-5'-oligoadenylate synthetase 2) | 71 | 1.88 |
| 8 | ISG15 (interferon-stimulated gene 15) | 71 | 3.42 |
| 10 | OAS1 (2'-5'-oligoadenylate synthetase 1) | 68 | 1.83 |
| 10 | IFIT3 (interferon-induced protein with tetratricopeptide repeats 3) | 68 | 3.46 |
4.3.1. Evaluation of Hub Gene in the GEPIA Database
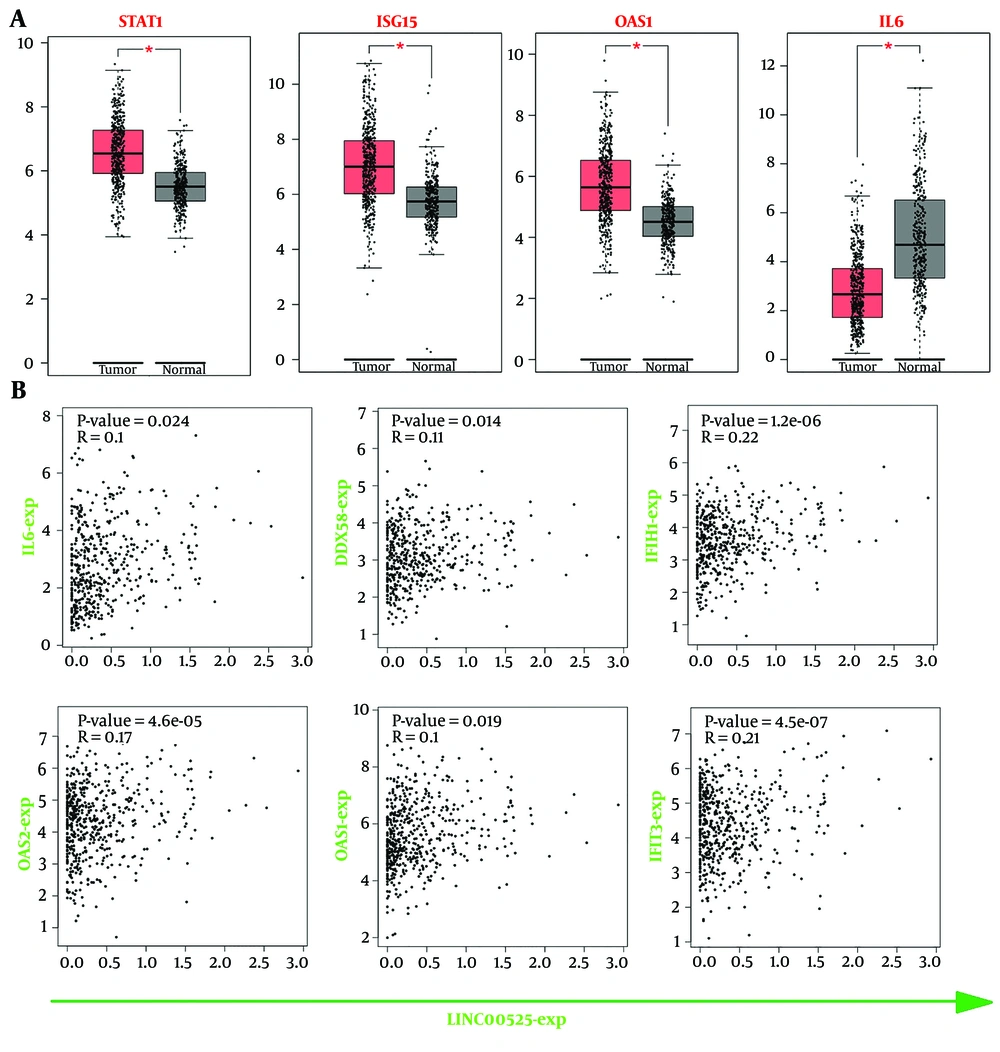
According to Pearson correlation analysis using the GEPIA online tool, mRNA expression levels of STAT1, ISG15, and OAS1 were significantly increased, while IL6 expression was decreased in LUAD tumor samples compared to normal samples (Figure 4A). Additionally, the GEPIA database showed that decreased expression of LINC00525 correlated with increased expression of IL6, DDX58, IFIH1, OAS1, OAS2, and IFIT3, which aligns with the results of this study (Figure 4B).
Evaluation of hub genes in GEPIA online website. A, differential expression of STAT1, ISG15, OAS1, and IL6 was significant in LUAD tumors compared to adjacent normal tissues according to the GEPIA; B, the expression of LINC00525 significantly is related to IL6, DDX58, IFIH1, OAS1, OAS2, and IFIT3.
4.4. Possible Effect of LINC00525 on mRNAs Through Interaction with Transcription Factors
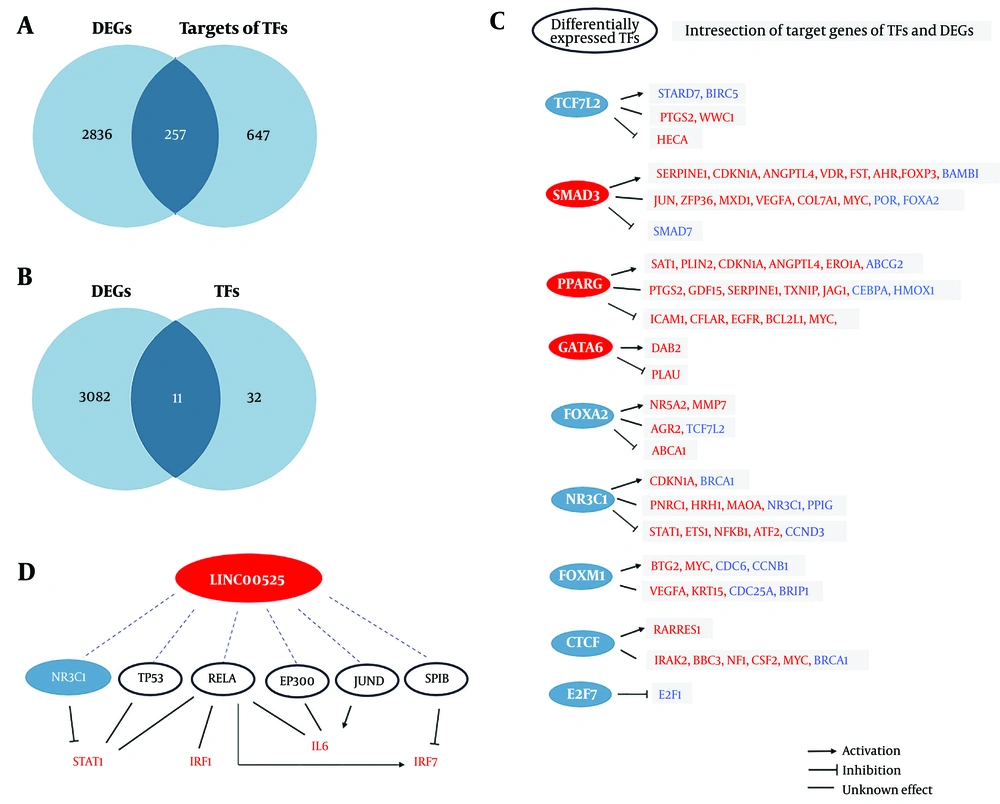
LncRNAs can activate or suppress the expression of downstream target genes through interactions with TFs. Based on the RNAInter database, 43 TFs that interact with LINC00525 were identified (score ≥ 0.3). The targets of these TFs were then identified using the TRRUST database. LINC00525 can influence the expression levels of these TF targets. A Venn diagram revealed 257 overlapping genes between DEGs and TF targets (Figure 5A). Notably, 11 TFs were also among the genes modified after LINC00525 knockdown, with some sharing common targets with DEGs (Figure 5B and C). Additionally, Figure 5D illustrates the TFs of hub genes that interact with LINC00525.
TFs influenced by LINC00525 and their regulatory networks based on RNA Inter and TRRUST databases. A, 257 DEGs were among the targets of affected TFs by LINC00525; B, 11 transcription factors (TFs) were identified as differentially expressed genes (DEGs); C, target DEGs that can be regulated by differentially expressed TFs. KDM5B gene did not exist in TRRUST database and FOXP1 has no common target genes with DEGs. Upregulated genes were written in red and downregulated ones in blue. Nods in red represent upregulated TFs and nodes in blue represent downregulated TFs; D, hub genes’ TFs which have interaction with LINC00525. Node in blue represents downregulated TF and white nodes represent unchanged TFs. Upregulated hub genes are written in red.
5. Discussion
Cell cycle progression has been reported as one tumorigenic function of overexpressed LINC00525 in LUAD (9). High expression of LINC00525 also leads to increased expression of cancer stem cell markers and chemoresistance in colorectal cancer cell lines (11). Moreover, the oncogenic role of LINC00525 has been documented in other cancers. For instance, LINC00525 promoted tumorigenesis and metastasis in spinal chordoma cell lines, while its inhibition enhanced apoptosis (12). LINC00525 has also been associated with survival and prognosis in oral squamous carcinoma, where its knockdown caused cell cycle arrest and apoptosis in carcinoma cells (12). The overexpression of LINC00525 across various cancers underscores its significance in tumorigenesis. Additionally, LINC00525 knockdown has been shown to induce various biological changes, including cell cycle arrest, apoptosis, drug sensitivity, and anti-invasive effects (8).
Given the regulatory effects of lncRNAs on gene expression, changes in lncRNA levels can modulate the transcriptome (7). Therefore, LINC00525 downregulation or upregulation can alter normal gene expression profiles, leading to changes in biological function and cell behavior (3). To date, several TFs, microRNAs, and mRNAs have been reported to be influenced by LINC00525. This study aimed to comprehensively assess the transcriptional changes in mRNAs following LINC00525 silencing in A549 LUAD cells. The silencing of LINC00525 caused significant changes in the mRNA profile of A549 LUAD cells, confirming the critical role of LINC00525 in gene expression regulation. As shown in Figure 2, LINC00525 silencing led to greater fold changes in upregulated genes compared to downregulated genes in A549 LUAD cells.
The cell cycle was identified as the primary cellular process associated with LINC00525 in A549 cells, as DEGs were significantly involved in this pathway. Accelerated cell division is a hallmark of cancer cells, often driven by mutations or disturbances in gene expression (13). Fang et al. demonstrated that the knockdown of LINC00525 led to G1 phase cell cycle arrest in A549 cells (9), and the data from this study further support this finding. Targeting cell cycle progression is a key therapeutic strategy for many anticancer drugs (13).
Cytokine signaling was another significantly enriched pathway among the DEGs. As shown in Figure 5, TFs of cytokine genes, including IRF1, IRF7, and IL6, could be influenced by LINC00525. Dysregulation of the cytokine regulatory network is a common feature in oncogenesis (14). Cytokines influence systemic inflammation and the immune tumor microenvironment, affecting various intracellular signaling pathways involved in tumor development. Cytokines can play both tumor-promoting and tumor-suppressing roles (15), and cytokine-based cancer therapies have been proposed (14). Dysregulation of cytokines is closely associated with cancer metastasis (16). In tumors, cytokines influence cancer cells and other components of the tumor microenvironment, leading to remodeling of the extracellular matrix, which can either support or hinder tumor growth and metastasis (17). This observation aligns with the positive association between LINC00525 overexpression and metastasis.
All identified hub genes were upregulated (Table 1), with many involved in the cytokine signaling pathway, including IL6, ISG15, IRF1, IRF7, IFIH1, and IFIT3. STAT1, another hub gene, belongs to the JAK-STAT pathway, a predominant signaling pathway activated by cytokines. These findings further confirm the role of the cytokine regulatory network in cell proliferation and tumor growth. Pearson correlation analysis also confirmed a correlation between the downregulation of LINC00525 and the upregulation of IL6, DDX58, IFIH1, OAS1, OAS2, and IFIT3. According to GEPIA, mRNA expression levels of STAT1, ISG15, and OAS1 were significantly increased, while IL6 expression was decreased in LUAD tumor samples compared to normal samples (Figure 4).
Silencing of LINC00525 in A549 cells led to the upregulation of all the aforementioned genes. STAT1, a transcription factor, plays key roles in various cellular biological processes, though its functions in tumor growth are not yet fully understood (18). One study reported that overexpressed STAT1 significantly inhibits the growth and invasion of lung cancer cells (19). Qu et al. demonstrated that upregulation of ISG15 inhibited tumor progression and migration of LUAD cells in both in vitro and in vivo experiments (20, 21). Although silencing LINC00525 led to a significant increase in OAS1, studies have shown that OAS1 promotes the proliferative activity of A549 LUAD cells (22, 23).
Regarding IL6, it has been reported that its activation of STAT3 results in cytotoxic CD8 T cell tumor infiltration, exerting a tumor-suppressing function. On the other hand, IL6 has also been found to promote tumors by activating cyclin D in LUAD (24). The tumor context plays a key role in determining IL6's functional direction, and it can have a preventive effect during the initiation of lung tumors (25). In this study, LINC00525 knockdown led to an increase in IL6 expression.
5.1. Conclusions
LINC00525 downregulation caused significant changes in the transcriptome of A549 LUAD cells. The evaluation of these altered genes provides insight into the molecular mechanisms by which LINC00525 promotes tumor growth and metastasis in LUAD. The most significantly enriched pathways for DEGs were the cell cycle and cytokine signaling pathways, both of which are commonly disrupted in cancers. Another key finding of this study was the identification of cytokine signaling-related hub genes, including STAT1, IL6, ISG15, IRF1, IRF7, IFIH1, and IFIT3. Further research is needed to explore other functional mechanisms of LINC00525, such as its interaction with differentially expressed microRNAs in A549 LUAD cells.