1. Background
Colorectal cancer (CRC) is one of the leading causes of cancer-related deaths worldwide, accounting for approximately 10% of all cancer cases (1). This disease poses a significant threat to millions of people globally, with metastatic disease occurring in around 20% of patients (2). At the molecular level, colorectal carcinogenesis involves the activation of oncogenes and the inactivation of tumor suppressor genes (3). Genes with highly differential expression patterns in CRC can serve as molecular markers, complementing histopathological factors in diagnosis and guiding therapeutic strategies for personalized patient care (4).
Noncoding RNAs (ncRNAs) are functional RNA molecules that are not translated into proteins, comprising 98% of the human genome (5). Long noncoding RNAs (lncRNAs) are a subclass of ncRNAs with more than 200 nucleotides and are evolutionarily conserved (6). Advances in understanding the molecular mechanisms of lncRNAs have shown that these molecules can function as either tumor suppressors or promoters in carcinogenesis (7). Abnormal lncRNA expression has been reported in various solid tumors (8, 9). As a result, lncRNAs are emerging as attractive targets for therapeutic interventions against cancer and as potential diagnostic and prognostic biomarkers in tumors (10-12).
Bacteria have the ability to target both primary tumors and metastases, making them useful for tumor-selective drug delivery (13-15). Escherichia coli K-12 is a well-known facultative anaerobic bacterium and human intestinal commensal (16). Due to its increased abundance in necrotic areas rather than the peripheral proliferative regions of tumors, the hypoxic areas of solid tumors could be targeted for treatments using this bacterium (17). Although E. coli K-12 has been reported to inhibit CRC progression through altered expression of protein-coding transcripts (18), no studies have yet examined its influence on ncRNAs. Given the role of oncogenic lncRNAs in cancer development, we aimed to investigate the impact of E. coli K-12 on downregulating the lncRNA MIR17HG.
In the present study, we evaluate the potential of E. coli K-12 to reduce MIR17HG expression, an oncogenic lncRNA with high expression in CRC patients. Furthermore, the Cox regression test was used to assess the effect of MIR17HG on CRC patient survival.
2. Objectives
This study aims to investigate the impact of E. coli K-12 on reducing CRC malignancy by altering MIR17HG expression. To achieve this, data from the cancer genome Atlas (TCGA) and the GEO database were used to identify the relationship between MIR17HG and E. coli K-12 in CRC. Additionally, this study assesses the effect of MIR17HG on CRC patient mortality through Cox regression analysis.
3. Methods
3.1. Data Sources, Normalization, and Differential Expression
To identify noncoding RNAs (ncRNAs) associated with the development and malignancy of CRC, transcriptomic data (RNAseq) was obtained from the TCGA database. For this purpose, the raw format (HTseq-Counts) of CRC RNAseq data was downloaded using the TCGAbiolinks package (19). The CPM (count per million) criterion of less than 10 was used to filter out genes with zero or near-zero expression using the edgeR package (20). The data were then normalized by the TMM (trimmed mean of M values) method and transformed into logarithmic form (base 2) using the limma package and the voom method (21). Subsequent analyses were conducted using the normalized expression matrix.
The cancer genome Atlas data were divided into cancer and normal groups, and the differential expression between these two groups was calculated using the linear model method. To investigate the relationship between the candidate lncRNA and E. coli K-12 in CRC, the GSE50040 dataset was analyzed from the GEO database. The raw expression profile data related to the study were downloaded and processed using the Affy package. Afterward, data normalization and logarithmic transformation (base 2) were carried out using the limma package and the RMA method (22). Samples from GSE50040 were divided into two groups, and the linear model method was applied to calculate expression differences between the treatment group with E. coli K-12 and the control group.
3.2. Clinical Data Processing and Survival Testing
The latest update of CRC clinical data was downloaded from the TCGA database to examine the role of MIR17HG in predicting patient survival. The normalized expression matrix was merged with the clinical data to assess the association between MIR17HG expression and patient prognosis using the Z-score scale. A Cox regression test was then performed to calculate the relationship between the expression level of MIR17HG and the patients' mortality rate. Finally, the Kaplan-Meier (K-M) curve was used to validate the results, with the median expression of MIR17HG set as the cut-off to divide the cancer samples into high and low expression groups.
3.3. Co-expression Network and Pathways
The RNAseq data from the TCGA database was utilized to investigate gene expression changes in CRC. Pathways related to MIR17HG were identified through the co-expression network and the normalized expression matrix. For this purpose, a Pearson correlation test was conducted between the expression level of MIR17HG and all genes in the expression matrix. Genes with R > 0.5 and P < 0.01 were selected as co-expressed genes. Pathways associated with these co-expressed genes were then identified using the MsigDB database through the Enrichr tool (https://maayanlab.cloud/Enrichr/).
3.4. Tissues Samples, RNA Isolation, cDNA Synthesis, and RT-qPCR
Forty CRC samples and an equal number of adjacent normal tissue samples were collected from the repository of the Iranian Tumor Bank. The review board of Imam Khomeini Hospital, where the samples originated, approved all bioethical aspects based on the regulations of the Iranian Ministry of Health, Treatment, and Medical Education. Ethical approval was granted under access number IR.IAU.PS.REC.1401.319. RNA extraction from the samples was performed using the TRIzol method (Sigma, USA) according to the manufacturer's instructions. cDNA synthesis was then conducted using the TaKaRa kit, following the company’s guidelines. To examine the expression level of the candidate lncRNA in CRC and adjacent normal tissues, specific primers for MIR17HG as the target gene and GAPDH as the internal control were designed using the Primer-Blast tool (NCBI). The primer sequences are summarized in Table 1. The expression level of MIR17HG was quantitatively assessed using the RT-qPCR method in both cancerous and normal tissue samples. The expression of MIR17HG was calculated using the 2-ΔCt method.
| Target Gene | Primer Name | Sequence |
|---|---|---|
| MIR17HG | Forward | 5’TCAGGAGTTCGAGACCAACC3’ |
| MIR17HG | Reverse | 5’TGCCTCAGCCTCCAGAGTAG3’ |
| GAPDH | Forward | 5’ACAGTCAGCCGCATCTTCT3’ |
| GAPDH | Reverse | 5’CCCAATACGACCAAATCC3’ |
The Specific Primer Sequences Used in This Study
3.5. Statistics and Software
All pre-processing, differential expression, co-expression network, and survival data analyses were conducted using the R programming language (v4.0.2), while GraphPad software (v8) was employed for generating and displaying graphs. The linear model method was applied to calculate the differences in gene expression between groups. The significance of the association between MIR17HG expression and patient prognosis was assessed using the log-rank test. Cytoscape (v4) was used to visualize the co-expression network of genes correlated with MIR17HG.
4. Results
4.1. Expression Changes of MIR17HG by Escherichia coli K-12 in CRC
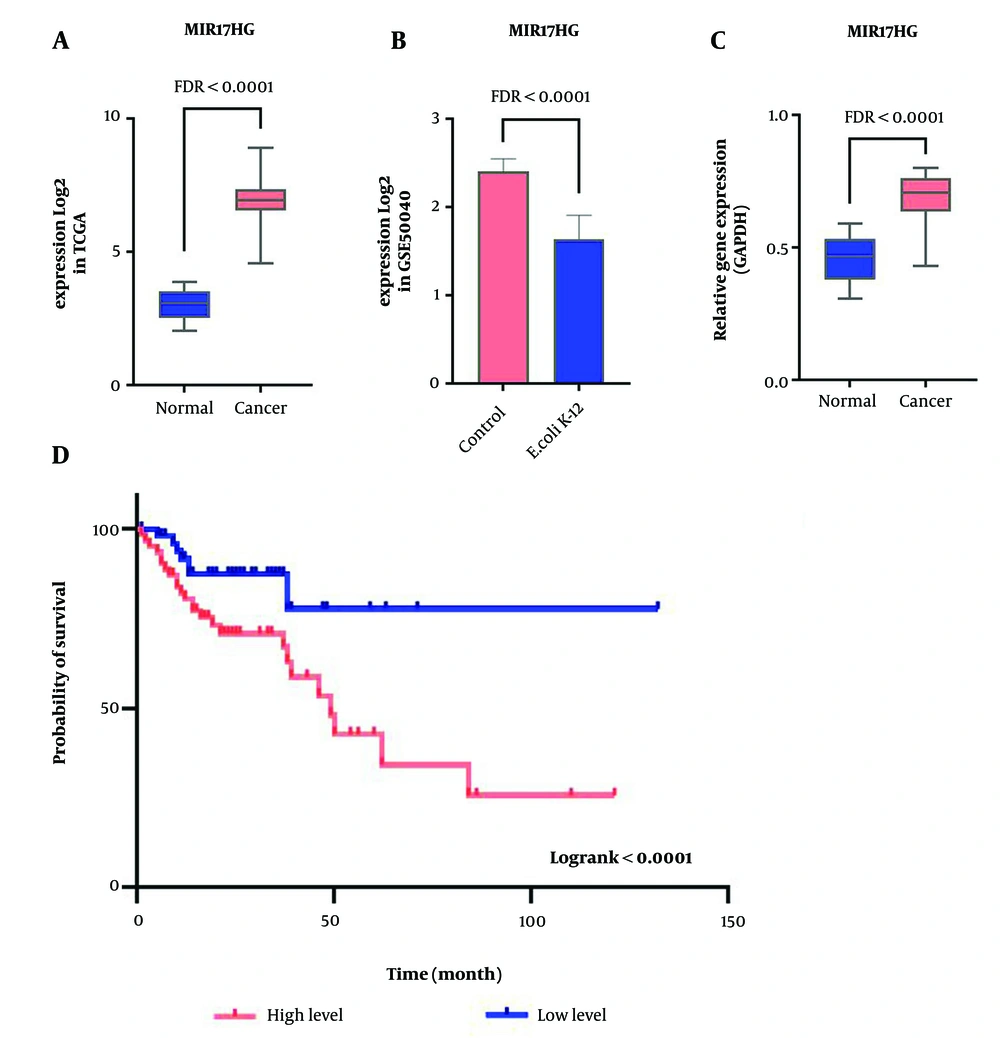
The lncRNA with significant expression changes under the influence of E. coli K-12 in CRC was identified based on the GSE50040 data analysis. Additionally, TCGA data was used to assess alterations in candidate lncRNA expression in CRC and normal samples. The analysis of expression differences revealed that the expression of MIR17HG was significantly increased in CRC samples compared to normal samples, with logFC > 1 and FDR < 0.01 (Figure 1A). In contrast, the expression of this lncRNA was downregulated in the presence of E. coli K-12, with logFC < -1 and FDR < 0.01 (Figure 1B). Furthermore, RT-qPCR results confirmed the in-silico findings, showing a significant increase in MIR17HG expression in CRC samples (Figure 1C). Enrichr analysis indicates that the dysregulation of mTOR, NF-kB, and BCR signaling pathways is highly associated with MIR17HG. These findings suggest that E. coli K-12 may impair tumor survival by reducing the overexpression of MIR17HG.
The high MIR17HG expression level as a poor prognosis biomarker downregulated by Escherichia coli K-12 in CRC. A, significant expression changes of MIR17HG in tumor samples compared to normal based on TCGA data; B, the E. coli K-12 impact on decreasing MIR17HG expression level; C, tthe RT-qPCR results for the expression of MIR17HG in forty CRC samples and adjacent normals; D, the association of MIR17HG expression with survival rate is shown through the Kaplan-Meier plot in CRC.
4.2. Relationship of MIR17HG and the Mortality Rate
The impact of E. coli K-12 on the candidate lncRNA and its association with malignancy and patient mortality was assessed by analyzing the relationship between MIR17HG expression levels and patient prognosis using TCGA clinical data. The Cox regression analysis revealed that MIR17HG (HR = 1.33, log-rank < 0.0001) is significantly associated with poor survival in CRC patients. Additionally, Kaplan-Meier analysis showed that increased MIR17HG expression is correlated with a higher mortality rate among patients (Figure 1D). These findings suggest that MIR17HG expression can serve as a prognostic biomarker in CRC.
4.3. Correlating of MIR17HG with Genes Related to the Main Pathways of Cancer
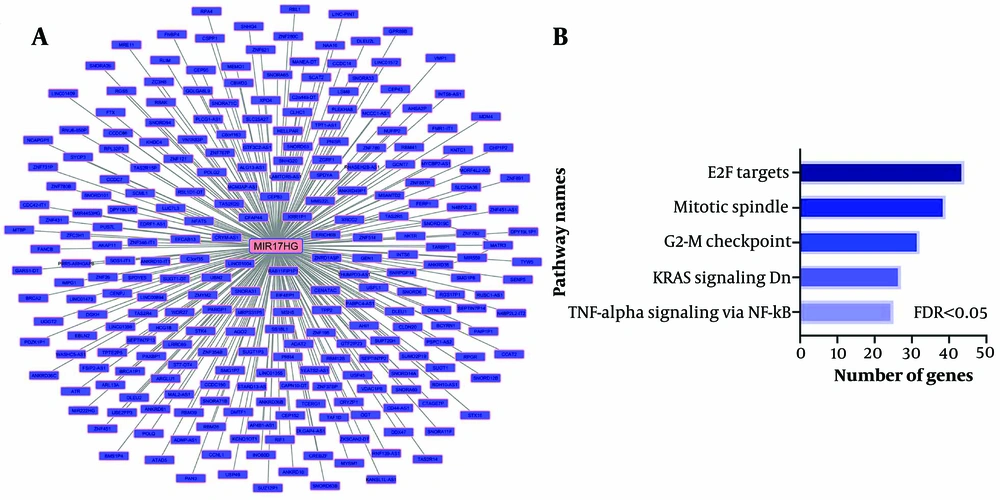
A co-expression network was used to identify pathways related to MIR17HG. The analysis revealed that MIR17HG expression levels are significantly correlated with 264 other genes, with a correlation coefficient greater than 0.5 and P < 0.01 (Figure 2A). These genes are involved in pathways such as TNF-alpha signaling via NF-kB, KRAS signaling, G2-M checkpoint, mitotic spindle, and E2F targets (Figure 2B, FDR < 0.01). These findings indicate that MIR17HG may contribute to the pathogenesis of CRC through these pathways.
Co-expression of MIR17HG with genes related to cancer development pathways. A, the co-expression network of all genes that had an expression correlation with MIR17HG at R > 0.5 and P < 0.01. The Pearson correlation test was performed between MIR17HG expression and all genes in TCGA colorectal cancer samples; B, enrichment results of all correlated genes with MIR17HG demonstrate their significant role in cancer progression pathways.
5. Discussion
Escherichia coli K-12 is an ideal anticancer bacterial strain due to its ability to specifically accumulate within tumors while being rapidly cleared from the rest of the body (13, 17). Recent evidence suggests that this bacterium can inhibit the development of CRC by affecting the transcriptome of cancer cells (18). In this study, we investigated the role of E. coli K-12 in altering the expression of MIR17HG, a key lncRNA in CRC carcinogenesis.
It has been reported that lncRNA MIR17HG plays a significant role in CRC liver metastasis (CRLM) by promoting aerobic glycolysis in CRC patients. MIR17HG transcription is elevated due to glycolysis-accelerated lactate accumulation via the p38/Elk-1 pathway (23). Our findings showed that the expression levels of MIR17HG in tumor samples significantly increased compared to normal tissues. Additionally, our survival analysis revealed that elevated MIR17HG expression is strongly associated with poor prognosis and CRC malignancy in patients. Furthermore, in silico results indicated the potential of E. coli K-12 in reducing MIR17HG expression in CRC cells.
The co-expression network in our study demonstrated that genes co-expressed with MIR17HG were associated with pathways such as TNF-alpha signaling via NF-kB, KRAS signaling, G2-M checkpoint, mitotic spindle, and E2F targets. Previous research shows that these pathways are related to cancer cell proliferation, spindle abnormalities, genomic instability, inhibition of apoptosis, cancer cell migration, invasion, and malignancy through their effects on immune cell infiltration (24-27). These findings suggest that MIR17HG plays a critical role in CRC pathogenesis through these pathways.
In summary, both in silico and ex vivo results of this study demonstrated that MIR17HG expression significantly increases in CRC, likely contributing to cancer development and malignancy. Additionally, we concluded that E. coli K-12 may modulate MIR17HG expression, potentially inhibiting cancer progression. However, further in vitro and in vivo studies are required to confirm the role of E. coli K-12 in reducing cancer progression by downregulating MIR17HG.
5.1. Conclusions
According to our findings, MIR17HG exhibits oncogenic properties and can serve as a biomarker for CRC prognosis. Moreover, E. coli K-12 may play an inhibitory role in CRC development by reducing MIR17HG expression. Therefore, E. coli K-12 could be beneficial in decreasing the malignancy of CRC through its impact on MIR17HG.