1. Background
Malassezia yeasts include a group of lipophilic and/or lipid-dependent, unipolar budding yeasts. The natural habitant of these yeasts is the skin of humans and other warm blooded animals (1, 2); they are found in 90% of healthy adult’s skin. They can change their saprophytic state and become pathogenic under the influence of predisposing factors, for example changes in the cutaneous microflora and/or alterations in host defenses (3). Yeasts of the Malassezia species cause pityriasis versicolor and are associated with pathogenesis of skin of disorders such as seborrheic dermatitis and atopic dermatitis, and recent information suggests their involvement in psoriasis (4). The first pityriasis versicolor disease was observed in 1801 by Willan. In 1889, Baillon suggested that the genus of Malassezia was the cause pityriasis versicolor (5, 6). In 1996 Gemmer et al. based on 28S rRNA gene sequences classified Malassezia genus to seven species (7). Fourteen species of Malassezia genus have been identified based on phenotypic features and molecular methods (8). The genus Malassezia includes the following species: Malassezia furfur, M. globosa, M. restricta, M. obtuse, M. sympodialis, M. slooffiae, M. pachydermatis, M. dermatis, M. japonica, M. nana, M. yamatoensis, M. equina, M. capra and M. cuniculi (3).
2. Objectives
The aim of the present study was to determine the frequency of common Malassezia species in patients with pityriasis versicolor and seborrheic dermatitis using of the nested PCR, in the city of Ahvaz.
3. Patients and Methods
In this study 85 clinical samples were collected from patients suspected of pityriasis versicolor and seborrheic dermatitis that had referred to our department of medical mycology. In addition some samples were also taken from patients referred to clinical health centers of Ahvaz. The samples of pityriasis versicolor were taken from each portion of the body that is more frequently affected by Malassezia and in seborrheic dermatitis from scalp scales. Malassezia DNA was extracted from scotch tape and scalp scales and major rDNA complex was amplified using the nested PCR assay with the general fungal ITS 1/4 and special primers.
3.1. Sample Collection
In patients with pityriasis versicolor, samples were collected by applying scotch tapes to lesional skin and in patients with seborrheic dermatitis, skin scales were scraped off by a sterile blade. A portion of the pathological samples was microscopically examined with methylene blue, which resulted in the appearance of many yeast cells and hyphae.
3.2. DNA Extraction
The Malassezia DNA was extracted according to methods of Sugita et al. (8). Briefly, scotch tapes samples of pityriasis versicolor and shells obtained from patients with seborrheic dermatitis were divided into small pieces and separately placed in a 2 mL eppendorf tube with 1.5 mL of lysis buffer (100 mM Tris-HCL [pH 8.0], 30 mM EDTA[pH 8.0], 0.05% sodium dodecyl sulfate) and incubated for 30 minutes at 100ºC. The scotch tapes were removed from the eppendorf tub and discarded. The suspension was extracted with 250 µL of phenol chloroform-isoamyl alcohol (25:24:1 vol/vol/vol). The samples were then extracted with 250 µL chloroform-isoamyl alcohol (24:1, vol/vol) and DNA was precipitated from the aqueous fraction with 2-propanol. Eventually the DNA pellet was washed in 70% cold ethanol and re-suspended in 50 µL of deionized water.
3.3. Detection of Malassezia by Nested PCR
Nested PCR was conducted using two sets of primers as shown in Table 1. In first round of PCR, ITS1/4 was used and in the second round of PCR, species-specific primers were used that were derived from the ITS region of the rRNA gene (8). Extracted DNA (10 µL) from each sample was added to 40 µL of the PCR master mix which consisted of 5 µL of 10 × PCR buffer (Cinnagen, Iran), 1.5 µL of 50 mM MgCL2 (Cinnagen, Iran), 1 µL of a 10 µM deoxynucleotide triphosphate mixture (an equimolar mixture of dATP, dCTP, dGTP and dTTP, Cinnagen, Iran), 0.3 µL of each primer (18S forward primer and 28S reverse primer), 0.5 µL of 5 U/µL Tag DNA polymerase (Cinnagen, Iran) and 34.1 µL of deionized water.
| Species and Primers Used | Sequence (5ʹ 3ʹ) | DNA Fragment Size, bp |
|---|---|---|
| First PCR for three Malassezia species | ||
| ITS1F-N | GGATCATTAGTGATTGCCTTTATA | |
| ITS4-R | TCCTCCGCTTATTGATATG | |
| Second PCR for each Malassezia species | ||
| M. furfur | 230 | |
| M. f-F | CTACTCGCGTACAACGTCTCTG | |
| 5.8S-R | TTCGCTGCGTTCTTCATCGA | |
| M. globosa | 270 | |
| M.gl-F | CAATAAGTGTGTCTCTGCGG | |
| 5.8S-R | TTCGCTGCGTTCTTCATCGA | |
| M. restricta | 320 | |
| M. rt-F | CTTGGTTGGACCGTCACTG | |
| M. rt-R | AGGCGGATGCAAAGTGTCTC |
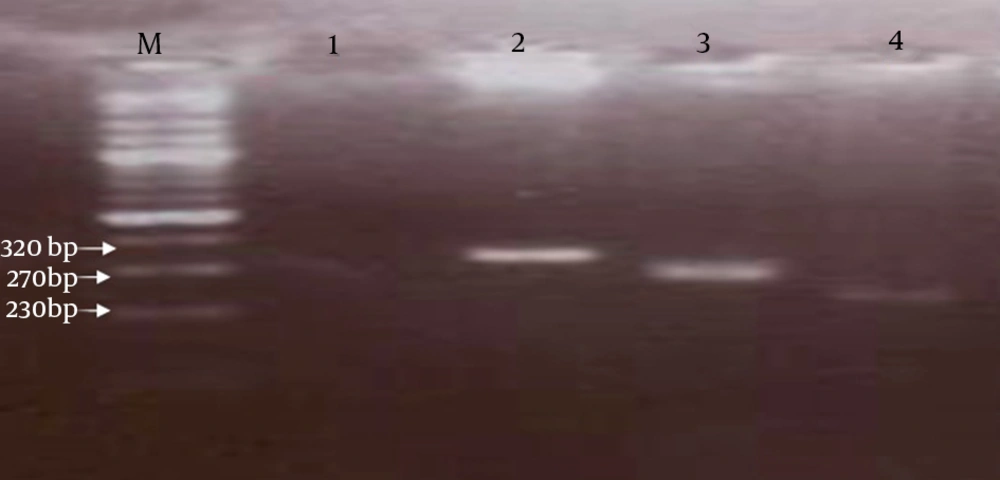
Amplification was performed in a thermocycler (BioRad, USA) with pre-denaturation for five minutes at 94ºC, followed by 30 cycles including 30 seconds at 94ºC, 1 minute at 57ºC and 50 seconds at 72ºC and a final extension for 10 minutes at 72ºC. During the nested PCR step, 1 µL of the first PCR product was added to a new reaction mixture consisting of the same components as the first PCR reaction mixture. The nested PCR procedure consisted of pre-denaturation at 94ºC for three minutes, followed by 30 cycles of 30 seconds at 94ºC, 60 seconds at 62ºC, 40 seconds at 72ºC and 10 minutes at 72ºC as the final extension. Nested PCR products were confirmed by electrophoresis on a 2% (wt/vol) agarose gel; stained with ethidium bromide using 0.5X tris-acetate-EDTA buffer and then visualized by the gel documentation system (Gel Doc, BioRad) (8, 9) (Figure 1).
4. Results
Among the 85 isolates of patients that were clinically suspected of pityriasis versicolor and seborrheic dermatitis, 75 (88.23%) isolates were stained by methylene blue, indicating that they were from the Malassezia genus. By using the nested PCR assay, three Malassezia species were detected at different rates in the samples of patients with seborrheic dermatitis and pityriasis versicolor. In this study, the most common species in pityriasis versicolor and seborrheic dermatitis were M. furfur (51.3%) and M. restricta (65.2%), respectively. Other species were M. globosa (35.2%) and M. restricta (13.5%) in pityriasis versicolor and M. globosa (26.1%), M. furfur (8.7%) in seborrheic dermatitis. In our study we didn’t detect any other species (Table 2).
| Malassezia species | Pityriasis Versicolor (n = 45) | Seborrheic Dermatitis (n = 30) |
|---|---|---|
| M. furfur | 19 (51.3) | 2 (8.7) |
| M. globosa | 13 (35.2) | 6 (26.1) |
| M. restricta | 5 (13.5) | 15 (65.2) |
| Malassezia spp. | 8 (17.8) | 7 (23.3) |
aData are presented as No. (%).
5. Discussion
The Malassezia genus belongs to the class of Ustilaginomycetes. Various species of the lipophilic yeast, Malassezia, are known as normal flora of the human skin and warm blooded animals, yet under certain conditions they can cause disease in humans and other animals, in which the seborrheic dermatitis and pityriasis versicolor disease are the most common (10). Since different Malassezia species respond differently to treatment, the disease caused by these agents doesn’t improve completely and recurrence is frequently observed. Therefore, for successful treatment, identifying the disease-causing agent is necessary. Methods for identifying these species include culture-dependent methods, such as physiological methods, and molecular methods (2, 10-12).
Using physiological methods has several disadvantages, for instance the high similarity in physiological test results between some species, such as M. furfur, M. sympodialis and M. slooffiae. Furthermore, in physiological methods, the need for specific environmental conditions, type of chemical materials and culture medium compounds limit the use of these methods for identify Malassezia species. Hence, it is essential to develop a sensitive method with high reproducibility that requires a small number of samples, to achieve correct identification of this yeast. Molecular methods are suitable for this purpose because these methods are very accurate, sensitive and rapid and can be used to identify organisms at the species level and even subspecies (13).
Methods based on PCR, such as single-step PCR, PCR-RFLP, ITS-rDNA regions sequencing, Real-time PCR, nested-PCR and genes encoding proteins are some of these methods (4, 8, 9, 14). The current study aimed to identify the dominant species in Ahvaz using the nested-PCR assay in patients with pityriasis versicolor and seborrheic dermatitis. In most studies around the world, the PCR-RFLP technique has been used to identify organisms isolated from culture medium (9, 10, 13, 15, 16), yet in this study for the first time we used of scotch tape technique for sampling of skin lesions and used nested-PCR assay to identify Malassezia species. In the nested-PCR technique specific primers with high sensitivity are used in the second phase. In our study this technique was performed, due to the low amount of yeast resulting in a low amount of DNA.
In this study, from 37 (82.22%) samples from patients with pityriasis versicolor, the most common species was M. furfur (51.3%) followed by M. globosa (35.2%) and M. restricta (13.5%), respectively. In most epidemiological studies in the world, M. globosa has been isolated more than any other species of Malassezia from patients with pityriasis versicolor (15, 17-21). In this study, M. furfur was the predominant species in patients, this difference with previous studies might be justified by Malassezia species dispersion in different geographic regions and also differences in methods used to identify the species. Morishita et al. during 2006 in Japan by using the nested-PCR assay, identified nine species of Malassezia where M. globosa and M. restricta (both 93.9%) were the most common (21); these results were different from that of our study. Shams et al. in Tehran during 2006 by using PCR-RFLP reported that M. globosa was the most common species isolated from samples (10). This result is in contrast with the results of our study.
One reason for this difference in results could be due to differences in the distribution of causing agents of pityriasis versicolor in Tehran and Ahvaz cities. Giusiano et al. in a study on patients with pityriasis versicolor in Argentina during the year 2010 reported M. sympodialis as the most common species (16). The results of this study confided with the results of our study and most other studies around the world. Since in that study the PCR-RFLP technique was used, perhaps the lack of consistent results with other studies, could be that some species are sensitive to changing of environmental conditions and some have low growth rates, such as M. globosa, M. obtuse and M. restricta. Mahmoudirad et al. performed a study in 2008 on 60 clinical isolates of pityriasis versicolor lesions using the PCR-RFLP method and reported that the most common species isolated was M. furfur (36.67%) followed by M. globosa (30%) (22). These results were consistent with the results of our study.
In most studies done around the world, the dominant species was M. restricta in patients with seborrheic dermatitis. Lim et al. (23) in 2008 using the nested-PCR, and Lee et al. in 2011 using the PCR-RFLP method, attempted to identify Malassezia species in patients with seborrheic dermatitis. In patients with seborrheic dermatitis, M. restricta (47%) and M. furfur (27%) were the dominant species (9). The results of the mentioned studies, was consistent with our results. In this study, we investigated the frequency of three species of M. furfur, M. globosa and M. restricta, due the commonality of these species in various studies done around the world on pityriasis versicolor and seborrheic dermatitis disease (8, 9, 16, 21). Other species were not surveyed due to their lower incidence in previous reports.
This study was not devoid of limitations, and limitations included a low sample size and also the fact that only three Malassezia species were considered. The importance of the present study was the application of the scotch tape as a method of sampling that was not used by any other relevant previous studies. In all previous studies, DNA extraction was performed using cultivated specimens or clinical samples were prepared using scraping shells. Furthermore there have been no previous studies on identification of Malassezia species in Ahvaz. The results can give an idea about disease causing species common in this region and since species have different sensitivities to antifungal drugs these results with further research could facilitate with choosing appropriate treatment options (13). The nested-PCR is a rapid and repeatable method for identification of important Malassezia species and should be used on more patients in the future. In conclusion, the most common agents of pityriasis versicolor and seborrheic dermatitis were M. furfur and M. restricta, respectively.