1. Background
Torque teno virus (TTV) is the first human circovirus that was isolated from Japanese patients with unknown hepatitis in late 1997. TTV belongs to the Circoviridae family (1, 2). TTV is a non-enveloped virus with a single stranded circular DNA genome, and TTV is resistant to killing (3). TTV has two open reading frames (ORF), ORF-1 and ORF-2 (4).The prevalence and genetic analysis of TTV were determined by using the primers of the UTR and ORF-1 regions (5), the 5’ UTR region that is located below the TATA box. The UTR is partially conserved among different genotypes and is more accurate for determining the prevalence of TTV. Using primers of 5’ UTR allows for prediction of more accurate prevalence. Through analysis of the N22 with the ORF-1 virus, the genotypes of the virus have been identified and differ by more than 30%. The high genetic diversity of the virus makes detection by PCR extremely difficult. A primer should be selected that has the ability to identify the maximum number of virus genotypes. A semi-nested PCR technique was used to replicate the virus (1). TTV has 40 genotypes from five major phylogenetic groups (6).
People who are exposed to blood and blood products can be co-infected with multiple strains of TTV (7). TTV is frequently separated from patients with acute or chronic hepatitis. TTV infection is common among blood donors, as well as patients with liver disease, including cirrhosis and cryptogenic disease, TTV was initially introduced as a new hepatitis agent (8). The high prevalence of TTV among those exposed to blood transfusions has been observed. It seems that the prevalence of TTV in hemodialysis patients who have received a kidney transplant is also high (9). In addition, TTV has been found in bile, liver and feces, and reproduction within the bone marrow has also been seen (10). The high potential for transmission of TTV through blood and blood products and high levels of TTV in the liver (8) create the necessity to investigate the prevalence of the virus in blood donor populations and people with hepatitis (1).
2. Objectives
Given the prevalence of hepatitis B and C patients in Ahvaz, we aimed to determine the prevalence of TTV in hepatitis patients in Ahvaz.
3. Patients and Methods
Our study was a cross -sectional study and included samples of 30 patients with hepatitis B and 30 patients with chronic hepatitis C, who were confirmed by the Danesh laboratory of Ahvaz. Samples were collected from January 2011 to June 2012. Patients with AIDS, cirrhosis and acute hepatitis B and C were excluded from the study. After matching for the age and sex of patients, blood samples from 30 healthy subjects to be used as controls were prepared. All samples were transferred to the department of medical virology. Serum samples were isolated and were stored at -70°C. TTV-DNA was extracted from the samples using a high pure viral nucleic acid kit (Roche, Germany). The DNA virus was replicated with a semi-nested PCR method using the primers 5’-UTR. After blast to NCBI, takapouzist made primers. NG054 and NG147 primers were used for the first round of PCR reaction, and NG054 and NG132 primers were used for the second round of PCR reaction (1). Primer sequences are shown in Table 1.
| Primer Name | Detected Region | Primer Sequence |
|---|---|---|
| NG054 | 5’ UTR | 5′- TTT GCT ACG TCA CTA ACC AC -3′ |
| NG147 | 5’ UTR | 5′- GCC AGT CCC GAG CCC GAA TTG CC -3′ |
| NG132 | 5’ UTR | 5′- AGC CCG AAT TGC CCC TTG AC -3′ |
Primer Sequences
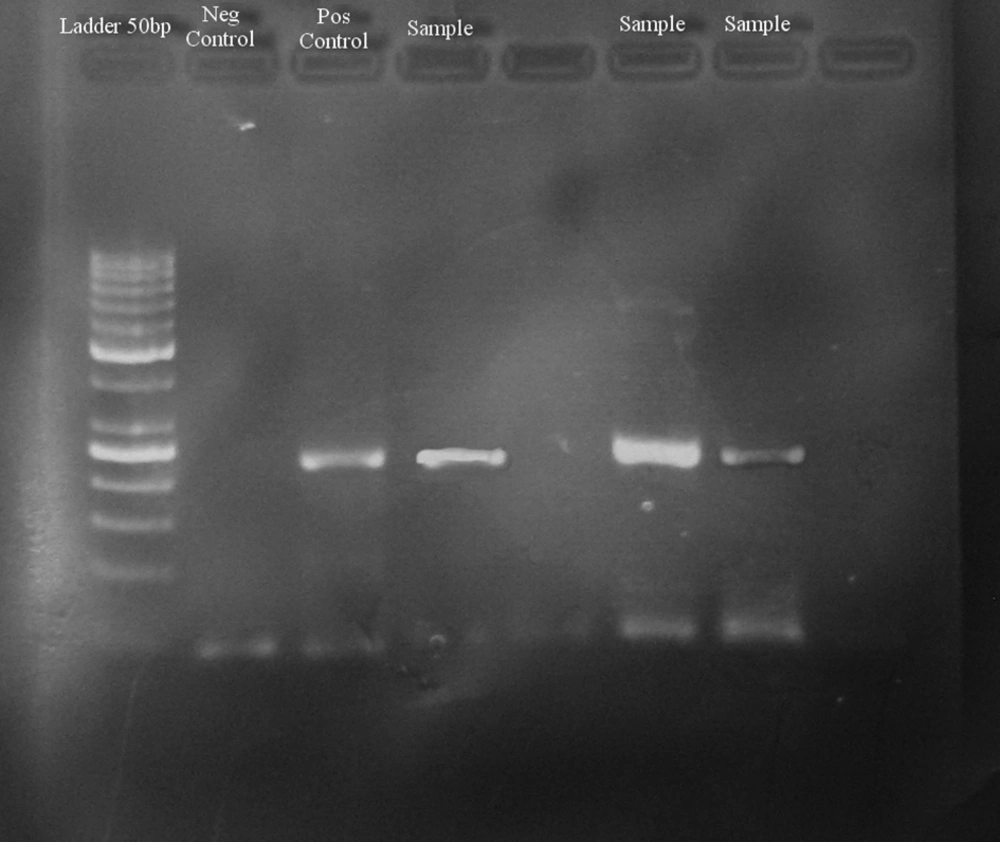
The first round of PCR was performed in 25 µL mixture, containing 0.25 µL of NG054 primer, 0.25 μL of NG147 primer, 2.5 μL 10x PCR buffer, 0.5 μL dNTPs, 10 µL template, 0.2 μL Taq DNA polymerase, 10.8 μL PCR water and 0.5 μL MgCl2. Also, PCR water was used as a negative control. A positive control was prepared from the blood transfusion center in Tehran, Iran. PCR thermal cycling conditions consisted of 94°C for 5 minutes 1 cycle. 94°C for 1 minute, 60°C for 1 minute, 72°C for 1 minute 40 cycles and a final extension of 72°C for 10 minutes. After the first round of PCR, the first round PCR product was used as the DNA template for the second round. The second round of PCR was performed in 25 μL mixture containing 0.25 μL of NG054 primer, 0.25 μL of NG132 primer, 2.5 μL 10x PCR buffer, 0.5 μL dNTPs, 7 μL template, 0.2 μL Taq DNA polymerase, 13.8 μL PCR water and 0.5 μL MgCl2. The program of the second round of PCR was similar to the first round, however, the extension was at 55°C for 1 minute, and the number of cycles was reduced from 40 cycles to 35 cycles. At the end the PCR, the products (220) were electrophoresed in 2% agarose gel for 30 minutes at 100 voltage, stained with SafeStain and photographed under ultraviolet light. In addition, a positive control (blood transfusion center, Tehran, Iran), negative controls and a DNA 50 bp ladder as a DNA molecular weight size marker were used.
4. Results
Among 60 samples from patients with chronic hepatitis B and C, there were 40 males and 20 females. Among males, 24 patients had chronic hepatitis C, and 16 patients had chronic hepatitis B. Among females, six patients had chronic hepatitis C, and 14 patients had chronic hepatitis B. Healthy subjects consisted of 20 males and 10 females. TTV-DNA was detected in nine out of 90 (10%) samples. The image of the gel is shown in Figure 1.
In detail, the separation of the nine samples was: six samples were males and three samples were females; three samples belonged to healthy subjects, six samples belonged to hepatitis patients (four samples to patients with chronic hepatitis C and two samples to patients with chronic hepatitis B); four samples of chronic hepatitis C patients consisted of one female in the age range of 50 - 55 and three males in the age ranges of 20 - 25, 30 - 35 and 40 - 45. The two samples of chronic hepatitis B patients consisted of one female and one male and belonged to the age range of 25 - 30 and 45 - 50, respectively. The three samples of healthy subjects consisted of one female in the age range of 45 - 50 and two males in the age ranges of 15 - 20 and 20 - 25.
The prevalence of TTV among chronic hepatitis B and C patients was (10%). The prevalence of TTV among chronic hepatitis B patients was (6.67%). The prevalence of TTV among chronic hepatitis C patients was (13.3%), and in healthy subjects was (10%). The prevalence of TTV among the general population was (10%). The number of patients and the prevalence of TTV in the age ranges is shown in Table 2.
| Age range | Patient | Healthy Subjects | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | Sex | HBV | HCV | TTV | Number | Sex | TTV | |||||||||
| Female | Male | Female | Male | Female | Male | HBV in Female | HCV in Female | HBV in Male | HCV in Male | Female | Male | Female | Male | |||
| 15 – 20 | 3 | 2 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 |
| 20 – 25 | 7 | 2 | 5 | 1 | 2 | 1 | 3 | 0 | 0 | 0 | 1 | 4 | 1 | 3 | 0 | 1 |
| 25 – 30 | 9 | 5 | 4 | 5 | 2 | 0 | 2 | 1 | 0 | 0 | 0 | 6 | 1 | 5 | 0 | 0 |
| 30 – 35 | 15 | 2 | 13 | 1 | 1 | 1 | 12 | 0 | 0 | 0 | 1 | 3 | 1 | 2 | 0 | 0 |
| 35 – 40 | 6 | 1 | 5 | 0 | 4 | 1 | 1 | 0 | 0 | 0 | 0 | 3 | 1 | 2 | 0 | 0 |
| 40 – 45 | 7 | 1 | 6 | 0 | 2 | 1 | 4 | 0 | 0 | 0 | 1 | 2 | 1 | 1 | 0 | 0 |
| 45 – 50 | 5 | 3 | 2 | 2 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 1 | 2 | 1 | 0 |
| 50 – 55 | 3 | 1 | 2 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 1 | 1 | 0 | 0 |
| 55 – 60 | 2 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 0 | 0 |
| 60 – 65 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 0 | 0 |
| 65 – 70 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| 70 – 75 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| Total | 60 | 20 | 40 | 14 | 16 | 6 | 24 | 1 | 1 | 1 | 3 | 30 | 10 | 20 | 1 | 2 |
Number of Patients and Prevalence of TTV in Age Rangesa
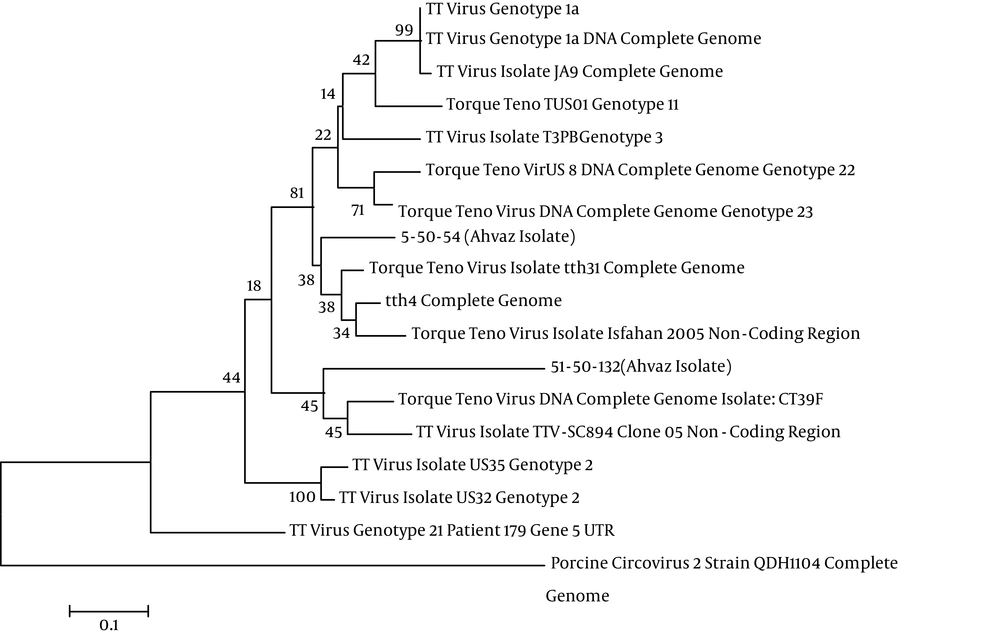
Two positive samples were sent to takapouzist for sequencing and both isolates, tth4 and tth31, were separated. Statistical analysis was conducted using the Chi-Square and t-test. P values of less than 0.05 were considered to be significant. However, in this study there was no significant association between positive TTV and age (P > 0.05) (Tables 3 and 4). Also, no significant association between positive TTV and sex was found (P > 0.05) (Tables 5 and 6).
Results of Independent Samples Test
| Value | df | Asymp. Sig. (2-sided) | Exact Sig. (2-sided) | Exact Sig. (1-sided) | |
|---|---|---|---|---|---|
| Pearson’s Chi-Square | 0.000a | 1 | 1.000 | ||
| Continuity Correctionb | 0.000 | 1 | 1.000 | ||
| Likelihood Ratio | 0.000 | 1 | 1.000 | ||
| Fisher’s Exact Test | 1.000 | 0.633 | |||
| N of Valid Cases | 90 |
Results of Chi-Square Tests
As seen in Tables 3 and 4, analysis showed an association between age and positive TTV. The t-test was used to indicate the association between age and positive TTV. Age was considered the dependent variable and the independent variable was positive TTV. The number of samples, mean, standard deviation and standard error for the dependent variable (age) are shown separately, and the TTV variable is defined for two groups. The number one indicates a positive TTV and the number zero indicates a negative TTV. Also, Levene’s test for equality of variances is shown. The results of the t-test for equality of means are shown in Table 4. The information that is reported on the equal variances line was not assumed for the t-test results when variances were not equal. The values for t, degrees of freedom (df) and p are also reported. In addition, the mean difference and standard error difference, as well as the 95% confidence interval of the difference are shown. Equal variances assumed tests often are used, because it is assumed variances are equal, A variance to the overall variance used for most accurate estimate of the common variance obtained. The P value is 0.486, showing that there was no significant association between positive TTV and age.
In Tables 5 and 6, the association between sex and positive TTV, as determined through analysis, is shown. We used the Chi-Square test to indicate the association between the two independent variables. The number one indicates a positive TTV and the number zero indicates a negative TTV. Also, F indicates female and M indicates male patients. The P value in this case indicated that there was no significant association between positive TTV and sex (P > 1).
5. Discussion
Previous epidemiological studies have been unable to estimate the true prevalence of TTV. Because of this, there is not a full understanding of the immune response generated by the presence of the virus in the host (11). Different prevalence rates of TTV have been reported in blood donors of different countries. For example, rates of 62% of blood donors in Italy (12), 28% of blood donors in China (13) and 42% of blood donors in Iceland (14) have been reported. In addition, the reports of the prevalence of TTV can even vary in different areas of a country, which indicates that the prevalence of TTV is associated to features of the genome (3, 11).
This study was performed to determine the prevalence and genotypes of TTV in patients with chronic hepatitis B and C and healthy subjects. In this study, primers from the 5' UTR region were used. The prevalence of TTV among chronic hepatitis B and C patients was 10%, and the prevalence of TTV among chronic hepatitis B patients was 6.67%. The prevalence of TTV among chronic hepatitis C was 13.3% and in healthy subjects was 10%. A study from Saudi Arabia by Almozaini et al., using 5’ UTR primers, on blood donors and hepatitis patients reported that DNA of TTV was detected in 101 (50.5%) out of 200 healthy blood donors. Also, DNA of TTV was detected in 39 (88.8%) out of 45 patients with chronic hepatitis and in 70 (70%) out of 100 patients with chronic hepatitis. Also, they detected a prevalence of TTV in 7 (12.5%). out of 56 non-A-G hepatitis patients (15). These results differ from the results of this study. This difference may be due to the geographical distribution of TTV, heterogenecity and the variability of the genome (16, 17).
Another study from India by Asim et al., using 5’ UTR primers, reported TTV-DNA was detected in 20 (21.5%) out of 93 samples from patients with chronic hepatitis, including chronic hepatitis B and C patients, hepatitis G patients, and non-A-G hepatitis patients. In this study, TTV-DNA was detected in 10.8%, 7.5%, 1.1% and 2.2% of patients with chronic hepatitis B, chronic hepatitis C, hepatitis G and non-A-G hepatitis patients, respectively (8). These results are similar to the results of this study. The similar results in both studies are justified, because the same primers were used and the number of samples was similar.
In another study in India by Irshad et al. on chronic hepatitis patients and healthy subjects, N22 primers were used and TTV-DNA was detected in 26 patients with chronic hepatitis (23.4%) out of 111 and in 27 (27%) out of 100 healthy subjects (6). Given that the primers used in both studies in India were different, the expected prevalence of TTV using the 5' UTR primers was higher than the prevalence of TTV using the N22 primers.
The genome of TTV is classified into 40 genotypes (2). The most common global genotypes of TTV are 1, 2 (18). The predominance of genotype 1a was observed in north India (8), while the most common genotype in hepatitis patients and healthy subjects in Saudi Arabia is 2c (15). The most common genotype of TTV in hepatitis patients in Iran is genotype 11 (1).
Two positive samples were sent to takapouzist for sequencing and both the tth4 and tth31isolates were separated. Nucleotides were aligned in NCBI and the same was done with the strains of TTV, however, the genotypes of the isolates were unknown. In Figure 2 the phylogenetic tree and isolates of TTV are depicted by the use of Mega5. The high prevalence of TTV is observed by using 5’ UTR primers in hepatitis patients and healthy subjects. This study did not obtain a high prevalence of TTV, perhaps due to:
1. TTV is a DNA virus, but its genetic diversity is increasing. The PCR methods used were not efficient enough, and they were not able to identify the types of TTV in blood (12).
2. Diagnosis of TTV by PCR depends on viral load. If the number of copies of the virus is low, negative test results are likely (18).
3. Some patients, at the time of sampling, received medications or treatment (e.g., ribavirin and interferon) (19, 20).