1. Background
Insulin is the most important hormone that regulates blood glucose levels in animals. Insulin malfunction leads to serious metabolic disturbances of diabetes mellitus (1). In human, the hormone is comprised of 51 amino acids in two chains; the A chain residues 1 - 21 and the B chain residues 22 - 51. Native insulin has three alpha helices in its secondary structure; two in the A chain and one in the B chain. There is a hydrophobic core of non-polar residues primarily in B chain that is essential for correct folding of insulin (2). In insulin tertiary structure, there are three disulfide bonds, two interchain bridges are made between A7-B7 and A20-B19 and one intrachain bridges residues of A6 and A11 simultaneously (3). It should be remembered that these disulfide bonds take part in protein stability and functionality. However, any deletions in disulfide bonds deactivate the hormone (4, 5). Nevertheless, these disulfide bonds cannot protect insulin at high temperatures. Under physiological conditions, zinc-coordinated trimers of insulin make dimeric super structures while the absence of zinc insulin makes dimeric or tetrameric associations (6-8).
Protein misfolding and aggregation are the main causes of certain diseases such as Alzheimer, Huntington, Parkinson and type 2 diabetes (9, 10). To study the mechanisms underlying such diseases, non-infective proteins can be used as a model to aggregate experiments. Currently, insulin is widely used as a useful model in this field, both for in vivo and in vitro studies (11). Previous studies showed that insulin is susceptible to fibrillation at acidic pH, heating and agitation conditions with rate dependence on the type of acid used and oligomeric status, i e, high rate of monomeric form (12). It is also shown that at acidic pH and high temperatures, oligomeric forms of insulin dissociate into monomers, with higher tendency to aggregate (7, 13). Increase in beta sheets is a common index for protein aggregation which can be used to follow aggregation and fibrillation (14). Moreover, there are increasing data regarding insulin folding, assembly and dynamics that help the precise study of structural basis for this phenomenon. For example, coordinate data on protein data bank (PDB) for insulin aggregates comprise the most exciting data in this context (15).
2. Objectives
The current study aimed to investigate the effect of different temperatures and pH conditions on insulin misfolding. The misfolding of insulin can be the first phase of aggregation. The investigations on protein misfolding and aggregation can help to find more effective cures for diseases arising from misfolding.
3. Methods
To study the effect of different temperatures and pH on insulin structure, a full length coordinate structure of insulin was needed. There were 199 available coordinate structures for insulin in PDB site. Among these structures, a wild type structure with PDB ID 1XGL obtained by nuclear magnetic resonance (NMR) spectroscopy with native conformation was selected. The acidic pH and extreme temperatures are well known parameters that threaten protein stability and promote its misfolding. Molecular dynamic simulation experiments provide a reliable tool for this study. Accordingly, the study carried out molecular dynamic experiments on insulin coordinate structure at neutral and acidic pH in three temperatures of 0°C, 37°C and 47°C separately. Insulin molecule was placed in the center of rectangular boxes, with dimensions of 3.75 × 4.39 × 3.84 nanometers for each experiment. Then the boxes were filled with explicit water molecules of SPC/E to mimic the native aqueous conditions. Acidic pH of the system was set by aspartate and glutamate in non-ionized state and assigning positive charges for lysine, arginine and histidine. However, neutral pH was set by considering ionizable residues in their ionized forms. To parameterize the simulated systems for molecular dynamics (MD) calculations the force field of 53A5 in GROMACS 4.5.5 with double-precision was used constantly. System neutralization which is a prerequisite for simulation was checked by preprocessor engine of GROMACS package and balanced by adding appropriate counter ions using genion command in GROMACS package. Energy minimization of the study systems that guarantee the safe progression of simulation was set to lower 300 kJ/M. Then, a short bonds-restrained molecular dynamic of 500 ps was performed prior to the final molecular dynamic simulation for 50 ns at one atmosphere total pressure. Other molecular dynamic parameters were set as reported before (16). Several analyses were done after simulation, including root mean square deviation (RMSD), root mean square fluctuation (RMSF), Gyrate, solvent accessible surface (SAS) and hydrogen bonds.
Protein secondary structure alterations were induced by 0°C and acidic pH in the systems was analyzed on PDBsum (protein data bank summary) website (http://www.ebi.ac.uk). This website provides the attributed Ramachandran plots. The Ramachandran plots were analyzed by visual molecular dynamics (VMD) software to follow amino acids contribution in protein secondary structure and misfolding process. All calculations were performed in Excel and SPSS.
4. Results
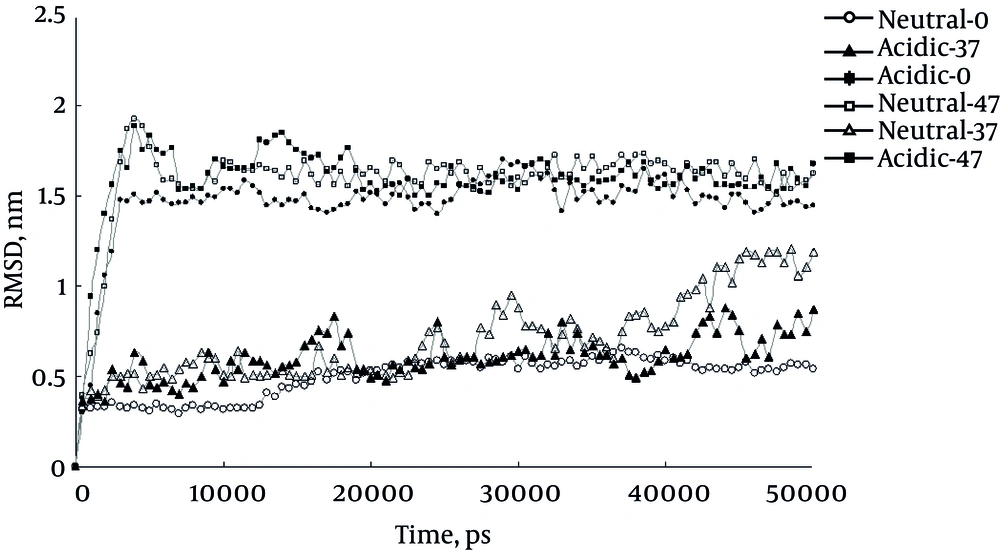
The present study carried out six MD simulations, three in acidic pH at temperatures of 0°C, 37°C and 47°C and three in neutral pH similarly. The first extracted result of the simulations was RMSD, shown in Figure 1. As it shows higher RMSD values for insulin at 0°C and acidic indicate more structural changes of insulin under these conditions. This finding means that acidifying the protein at 0°C induces these structural changes. However Figure 1 reveals that the same effect is not observed at 37°C. Increasing the temperature to 47°C masks these structural changes probably by increasing kinetic energy that accelerate conformational changes behind that excreted by pH. It should be mentioned that increase in RMSD curve could be interpreted as structural alteration, either in the direction of over folding or unfolding of protein.
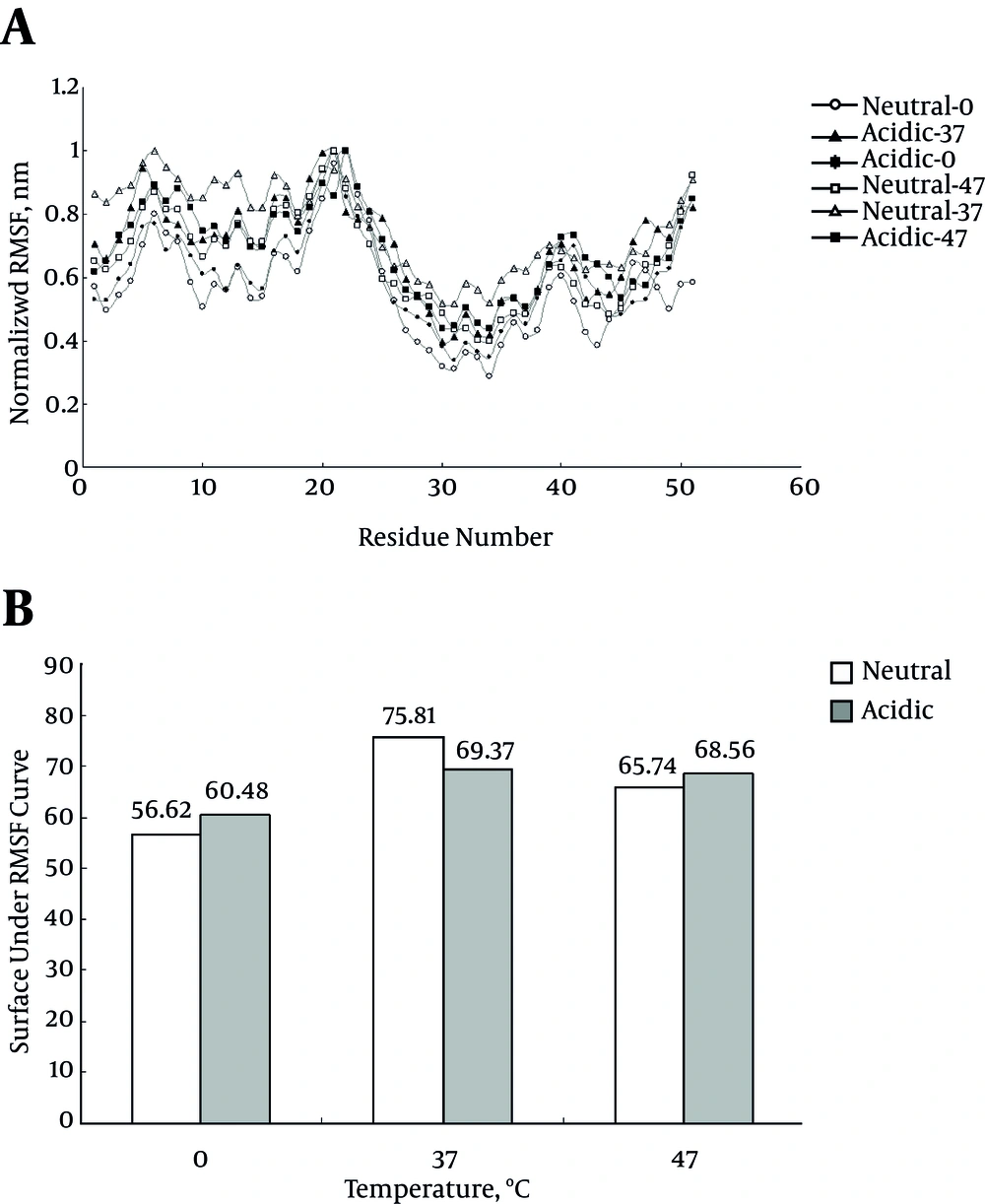
The RMSF for protein alpha carbons during simulation period comprises the useful index for protein flexibility at each residue. Figure 2A represents RMSF curve for six simulations. Since the curves do not have similar baselines, the comparison of RMSF curves is not applicable for the current study simulations. Therefore, the surface under each curve and represent the areas in Figure 2B was calculated. The larger surface under the curve is related to the more flexibility of protein and vice versa. Expectedly, the flexibility of protein at 0°C is significantly less than those of other temperatures; while acidic pH ameliorate this decreased flexibility at this temperature. The decrease in protein flexibility at 0°C is obviously related to decrease in kinetic energy when compared to other temperatures.
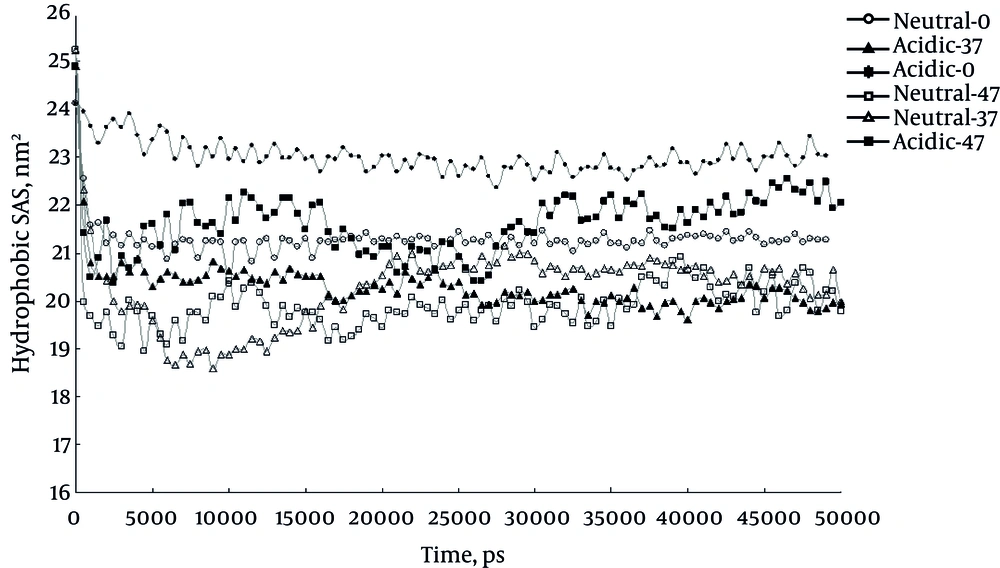
Figure 3 shows time course change in SAS for protein during simulation period. In confirmation to the current study results SAS curve shows higher values for protein at 0°C and acidic pH compared to other conditions. The elevated SAS could be interpreted as more structural changes.
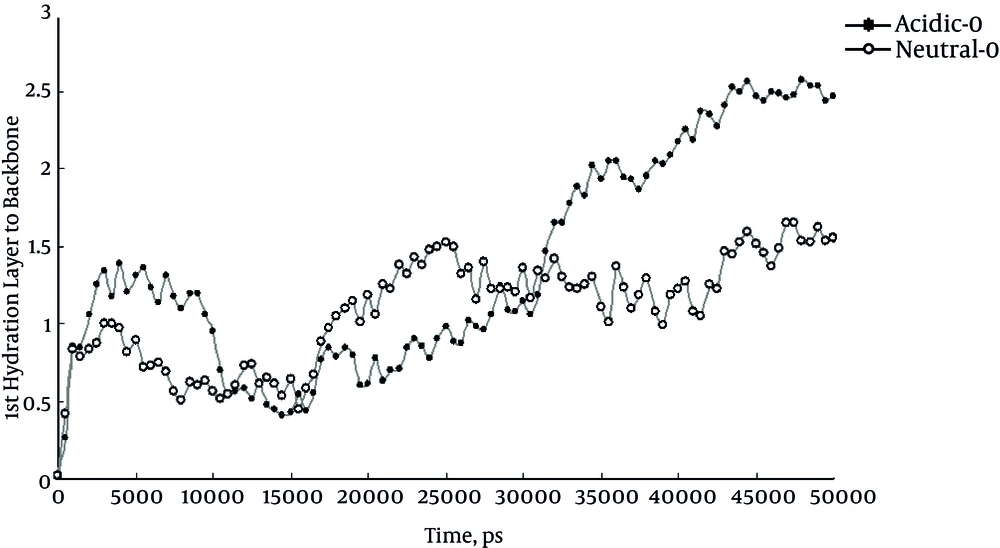
Another useful parameter to study the effect of acidity in this study was the distance between the first hydration layer and protein backbone or first hydration layer thickness. It is a useful index to follow with protein folding status. The increase of this distance known as desolvation phenomenon occurs while the protein is folding and vice versa. Figure 4 represents the distance between the first hydration layer and protein backbone at 0°C. Comparison of the curves shows the role of acidity in desolvation of insulin protein. But the same effect of acidic pH is not observed at 37°C and 47°C (data not shown).
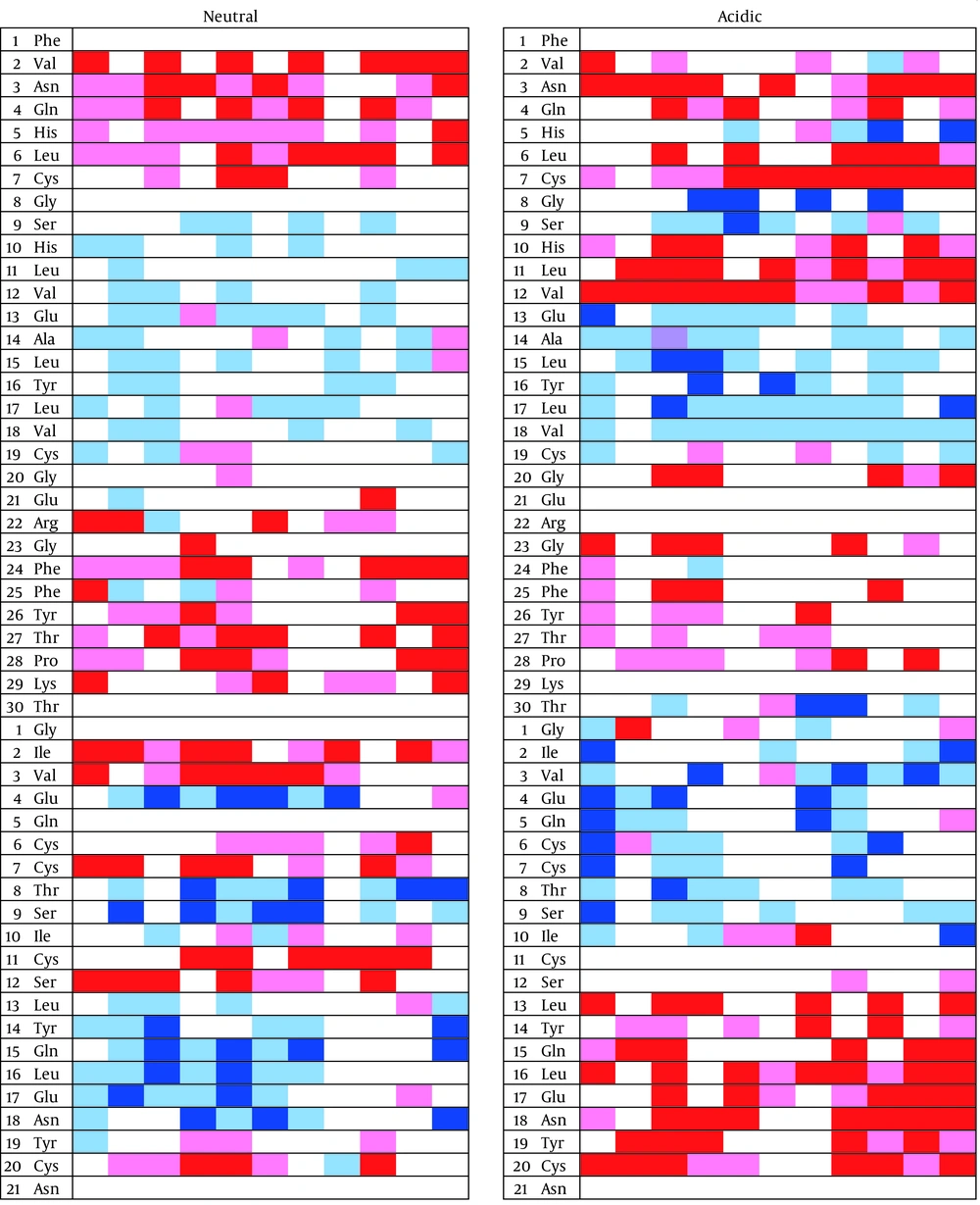
Secondary structure changes can provide valuable information about the mechanism of misfolding and aggregation. Figure 5 shows the status of each residue during the simulation at 0°C. There is an area between two residues Cys7 and Cys 19, surrounded by two disulfide bonds, that does not take any specific secondary structure at neutral pH. This unstructured state continues till the end of the simulation. Unlike acidic condition the area takes secondary structures and spends most of the simulation period in alpha helix or beta sheet structures. It should be noticed that acidifying the solution increases the beta sheet content in insulin structure.
Another important point is that the insulin peptide proceeds to gain more beta sheet structures. These beta sheets appear mostly in the B chain N-terminal and the A chain C-terminal.
Next column shows the times, each 5 ns. The red color represents beta sheet, pink almost allowed for beta sheet, dark blue alpha helix, and light blue for almost allowed for alpha helix. The non-colored regions represent the random coiled areas. Eventually Cys7 - Cys19 does not take any specific secondary structures. This area is surrounded by two disulfide bridges.
5. Discussion
Protein aggregation and misfolding are the underling phenomenon in serious diseases such as Alzheimer and type 2 diabetes. Insulin is one of the most used models in misfolding and aggregation studies. The present study simulated insulin structures at three temperatures of 0°C, 37°C and 47°C in neutral and acidic pH conditions to explore aggregation inducing agents. The results of the simulation are represented by molecular parameters. As reported before, analysis of the current study RMSD curves indicates the important role of low pH and temperatures as well as high temperatures on the structural changes during the simulation period (17). The study data indicate that at 0°C the acidic pH insulin shows higher RMSD values compared to that of neutral condition, probably because of higher structural alterations in such circumstances, the structural alterations are interpreted as protein misfolding and aggregations (18). Increased RMSD values and structural disturbance at high temperature of 47°C may be due to the high kinetic energy at this condition regardless of pH condition.
The misfolding process does not necessarily mean protein unfolding. Instead, misfolding may be manifested as protein partially unfolding or over folding through protein turnover in cells.
During misfolding event, one of the characteristics which changes during this process, is the protein flexibility degree. Protein flexibility index, RMSF, is a useful parameter to follow with the protein misfolding (19). Decreased RMSF at 0°C could be considered as decreased protein flexibility, which implies protein over folding. Expectedly, the flexibility of protein is significantly higher in acidic pH than neutral condition (P < 0.05) that can give protein the ability to experience more alterations that facilitates protein over folding. In fact the RMSD and RMSF results confirm the insulin over folding and misfolding simultaneously.
Another usable index in survey on misfolding is SAS area of the protein. The results of SAS parameter calculation in the current simulations indicate a significant increase at 0°C and in acidic condition, which expectedly confirms structural changes of protein that leads to misfolding.
As already reported, all calculated molecular parameters in the study confirmed the misfolding of insulin at 0°C and in acidic condition (20, 21). Unlike the study by Kachooei et al. the current study found that low temperature in presence of low pH can affect the insulin structure similarly or even more than high temperature (21). For more assurance, the study analyzed the thickness of first hydration layer. Increase of the distance between first hydration layer and backbone of protein shows that protein is misfolding, actually by over folding. To find out what regions are playing the critical role in over folding, analyses of the secondary structures were necessary. The region between residues Cys7 and Cys19 has the largest changes during acidification. The mentioned region that is surrounded by a disulfide bond can be a valuable point for more investigation about protein misfolding and aggregations, as well as the increase of total beta sheets.
The current study results confirmed several similar studies. Whittingham et al. worked on the conditions that promote the insulin misfolding. They confirmed the effect of acid and temperature on the insulin structure that can lead to misfolded aggregated insulin. One of the properties of the misfolded insulin was the increase in beta sheet structures (12).
Insulin is one the most studied proteins in biochemistry. Since 1922, after extraction, it became a popular molecule to study various factors (12). Recently, insulin became a popular model to study protein misfolding. The current study also used insulin as a suitable protein to investigate the effect of misfolding agents. Similar to the previous reports, the current study results showed the certain effect of temperature and acidity on protein misfolding.