1. Background
Nosocomial infections, also known as hospital-acquired infections, are common in burn patients due to the special features of the diseases, such as skin damage, physiological changes, prolonged hospitalization, and receiving aggressive interventions (1). Klebsiella pneumoniae belongs to the Enterobacteriaceae family and is described as a gram-negative, encapsulated, and lactose-fermenting bacteria (2) that is responsible for various hospital-acquired infections such as pneumonia, septicemia, diarrhea, liver abscess, endophthalmitis, meningitis, urinary tract infections, and bacteremia, whose mortality rates are high (3). Along with other gram-negative bacteria such as Pseudomonas aeruginosa and Acinetobacter baumannii, K. pneumonia pathogens are commonly isolated from patients with burn infections (4). Most of such infections are caused by multidrug-resistant (MDR) strains that interrupt the treatment processes (5). Besides, extended-spectrum beta-lactamase (ESBL)-producing K. pneumonia isolates are highly resistant to antibiotics, which further complicate infections and the treatment processes of burn patients. Moreover, MDR strains are resistant against beta-lactam antibiotics and different types of antibiotics, including quinolones and aminoglycosides (6). Quinolones and fluoroquinolones extensively use for the treatment of K. pneumonia-induced infections, mainly because of the resistance of these bacteria to the first-line drugs, which have led to a significant increase in resistance against them (7).
Plasmid-mediated quinolone resistance (PMQR) is mediated by the qnr genes, boosting antimicrobial resistance among bacteria due to their placement on mobile genetic elements. Therefore, PMQR also plays a significant role in quinolone resistance due to its high emission potential among Enterobacteriaceae (8). Three types of PMQR genes are identified so far, including the qnr, which is the most important one (9). In addition to resistance to quinolones, PMQR can be similarly effective in resistance against other antibiotics, especially beta-lactams and aminoglycosides (10). To date, five qnr groups are identified, including the qnrA, qnrB, qnrC, qnrD, and qnrS. Moreover, PMQR can exert its influence through two further mechanisms of QepA and OqxAB pumps and an aminoglycoside acetyltransferase known as the aac(6’)-Ib-cr, which cause reduced susceptibility against ciprofloxacin (9, 11, 12). The qnr genes also cause resistance against quinolones via inhibiting DNA gyrase and topoisomerase IV. Alongside aminoglycoside resistance, the aac(6’)-Ib-cr gene can correspondingly cause resistance against fluoroquinolones (13).
2. Objectives
No recent comprehensive research is performed on the prevalence rates of PMQR genes in K. pneumonia isolates among patients in the city of Kermanshah, Iran. Therefore, the present study aimed to determine the frequency of the qnrA, qnrB, qnrS, and aac(6’)-Ib-cr genes in K. pneumonia strains of burn wound infections collected from patients in the city of Kermanshah, Iran.
3. Methods
3.1. Isolate Collection and Recognition
The current descriptive-analytical study was conducted on 465 clinical samples collected from burn patients admitted to Imam Khomeini Hospital in Kermanshah from August 2017 to June 2018, who had burn wounds with no history of antibiotics consumption for more than a week. After collection, samples were immediately transferred to the laboratory under sterile conditions. Then, samples were cultured onto eosin methylene blue agar (EMB) and MacConkey agar (Merck Group, Germany) and subsequently incubated for 24 h at 37ºC. In total, 126 K. pneumonia isolates were detected using standard biochemical tests through culturing in triple sugar iron agar (TSI), sulfur-indole-motility (SIM), methyl red-Voges Proskauer (MRVP), citrate, and urea broths (3) (HiMedia Laboratories Pvt. Ltd.., India). Afterward, the identified K. pneumonia samples were maintained in trypticase soy broth (TSB) treated with 15% glycerol (-70°C).
3.2. Antibiotic Susceptibility Testing (AST)
The antimicrobial susceptibility patterns of samples were assessed via disk diffusion test based on the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (14) for ciprofloxacin (5 μg), levofloxacin (5 μg), enrofloxacin (10 μg), ofloxacin (5 μg), gatifloxacin (5 μg), nalidixic acid (30 μg), gentamicin (10 μg), amikacin (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), ceftriaxone (30 μg), and aztreonam (30 μg), as antibiotics provided by MAST (the United Kingdom). In cases that matching with the density of the 0.5 McFarland standards (1.5 × 108) was required, the concentration of the bacteria was used for antimicrobial susceptibility testing. The K. pneumonia, the American Type Culture Collection (ATCC) 700603, and the Escherichia coli, ATCC 25922 were further applied to ensure the quality of the antimicrobial susceptibility tests (3, 10).
3.3. ESBL Confirmation by Combination Disk (CD) Method
Isolates with a minimum inhibition zone diameter of 22, 25, and 27 mm, respectively, for ceftazidime, ceftriaxone, and cefotaxime were assessed for detecting ESBL genes. To confirm ESBL production, the CD method was further performed using 30 μg cefotaxime and ceftazidime disks impregnated with 10 μg clavulanic acid (MAST, the United Kingdom) on Mueller-Hinton agar (HiMedia Laboratories Pvt. Ltd., India) similar to the disc diffusion method. On the other hand, the strains with a minimum inhibition zone diameter equal to or more than 5 mm, in comparison with the single disc of the same antibiotic, were regarded as ESBL-producing (15).
3.4. Polymerase Chain Reaction (PCR)
After extracting the deoxyribonucleic acid (DNA) of the isolates by boiling, the frequency of the qnrA, qnrB, qnrS, and aac(6’)-Ib-cr genes was assessed using the PCR through special primers (Takapou Zist Co., Iran) (10), as shown in Table 1. The concentration of the DNA samples was determined at wavelength 260 nm using a NanoDrop Synergy HTX (Bio Tek Instrument, Inc Highland Park, USA) equal to 35 pmol/ul. Also, the purity of the extracted DNA at 260/280 nm was 1.85. PCR was accordingly performed using the total 25 μL volume, including Master mix (12.5 μL) (Sinoclon Co., Iran), 1 μL of the primers, bacterial DNA (2 μL), and sterilized distilled water until reaching 25 μL. The following stages were included in the PCR protocol: initial denaturation (94ºC/5 min), 30 main cycles according to Table 1, and extension (10 min/72ºC). The E. coli J53 strains, including pMG252, pMG298, and pMG306, were also considered, respectively, as positive controls of the qnrA, qnrB, and qnrS genes. Then, 1% agarose gel was applied for electrophoresis of the PCR yields, and the product was consequently stained by ethidium bromide.
| Primer | Sequence (5’-3’) | Denaturation, 94ºC | Annealing, 1 min | Extension, 72ºC | Product Size (bp) |
|---|---|---|---|---|---|
| qnrA | TTCTCACGCCAGGATTTGAG TGCCAGGCACAGATCTTGAC | 1 min | 57ºC | 1 min | 571 |
| qnrB | TGGCGAAAAAATTGAACAGAA GAGCAACGATCGCCTGGTAG | 1 min | 57ºC | 1 min | 594 |
| qnrS | GACGTGCTAACTTGCGTGAT AACACCTCGACTTAAGTCTGA | 1 min | 57ºC | 1 min | 388 |
| aac (6’)-Ib-cr | TTGCGATGCTCTATGAGTGGCTA CTCGAATGCCTGGCGTGTTT | 1 min | 54ºC | 1 min | 482 |
3.5. Statistical Analysis
Data were analyzed using the IBM SPSS Statistics version 20 via the chi-square test and Fisher’s exact test. As well, P values less than 0.05 were considered statistically significant.
4. Results
Analyzing 126 K. pneumonia isolates showed a prevalence of 73 (57.9%) and 53 (42.1%) for males and females, respectively. The mean age of participants was 36.11 ± 48.42, and the youngest and oldest participants were 11 and 73 years, respectively. Most of the isolates (44 or 34.9%) with K. pneumonia were taken from patients aged 31 to 45 years, and the lowest was from patients less than 6 years (6 or 4.8%). All isolates were obtained from patients with burn wounds admitted to Imam Khomeini Hospital in the city of Kermanshah (Iran). The prevalence rate of MDR isolates was 54.8% (69 isolates). Based on the findings of the phenotypic test, 52 (41.3%) isolates (out of the 126) were ESBL-producing. In the positive ESBL isolates, the frequency of MDR isolates was 46 (88.5%). According to Table 2, the ESBL-producing isolates indicated more antimicrobial resistance than non-ESBL-producing samples (P < 0.05). The highest antimicrobial resistance level in ESBL-producing and non-ESBL-producing K. pneumonia isolates was against ceftriaxone (90.4%) and nalidixic acid (47.3%), respectively. In addition, the highest level of susceptibility in ESBL-producing and non-ESBL-producing samples was, respectively, against gatifloxacin (40.4%) and amikacin (10.9%) (Table 2).
| Antibiotics | All Strains (N = 126) | ESBL Positive (N = 52) | ESBL Negative (N = 74) | P Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R | S | I | R | S | I | R | S | I | ||
| NA | 69 (54.8) | 51 (40.5) | 6 (4.8) | 34 (65.4) | 16 (30.8) | 2 (3.8) | 35 (47.3) | 35 (47.3) | 4 (5.4) | 0.108 |
| CIP | 66 (52.4) | 52 (41.3) | 8 (6.3) | 31 (59.6) | 16 (30.8) | 5 (9.6) | 35 (47.3) | 36 (48.6) | 3 (4.1) | 0.030b |
| LEV | 43 (34.1) | 76 (60.3) | 7 (5.6) | 23 (44.3) | 29 (55.7) | 0 | 20 (27) | 47 (63.5) | 7 (9.5) | 0.875 |
| OFX | 48 (38.1) | 72 (57.1) | 6 (4.8) | 24 (46.2) | 26 (50) | 2 (3.8) | 24 (32.4) | 46 (62.2) | 4 (5.4) | 0.320 |
| NOR | 59 (46.8) | 58 (46) | 9 (7.1) | 27 (52) | 23 (44.3) | 2 (3.8) | 32 (43.2) | 35 (47.3) | 7 (9.5) | 0.820 |
| GAT | 38 (30.2) | 88 (69.8) | 0 | 21 (40.4) | 31 (59.6) | 0 | 17 (23) | 57 (77) | 0 | 0.029b |
| AK | 42 (33.3) | 82 (65.1) | 2 (1.6) | 34 (65.4) | 17 (32.7) | 1 (1.9) | 8 (10.9) | 65 (87.8) | 1 (1.3) | 0.0001b |
| GM | 52 (41.3) | 72 (57.1) | 2 (1.6) | 37 (71.2) | 14 (26.9) | 1 (1.9) | 15 (20.3) | 58 (78.4) | 1 (1.3) | 0.0001b |
| CTX | 57 (45.2) | 69 (54.8) | 0 | 45 (86.5) | 7(13.5) | 0 | 12 (16.2) | 62 (83.8) | 0 | 0.0001b |
| CAZ | 52 (41.3) | 72 (57.1) | 2 (1.6) | 43 (82.7) | 19 (17.3) | 0 | 9 (12.2) | 63 (85.1) | 0 | 0.0001b |
| CRO | 61 (48.4) | 65 (51.6) | 0 | 47 (90.4) | 5(9.6) | 0 | 14 (19) | 60 (81) | 0 | 0.0001b |
| ATM | 51 (40.5) | 75 (59.5) | 0 | 42 (80.8) | 10 (19.2) | 0 | 9 (12.2) | 65 (87.8) | 0 | 0.0001b |
Abbreviations: AK, Amikacin; ATM, Aztreonam; CAZ, Ceftazidime; CIP, Ciprofloxacin; CRO, Ceftriaxone; CTX, Cefotaxime; GAT, Gatifloxacin; GM, Gentamicin; LEV, Levofloxacin; NA, Nalidixic Acid; NOR, Norfloxacin; OFX, Ofloxacin
aValues are expressed as No. (%).
bSignificant
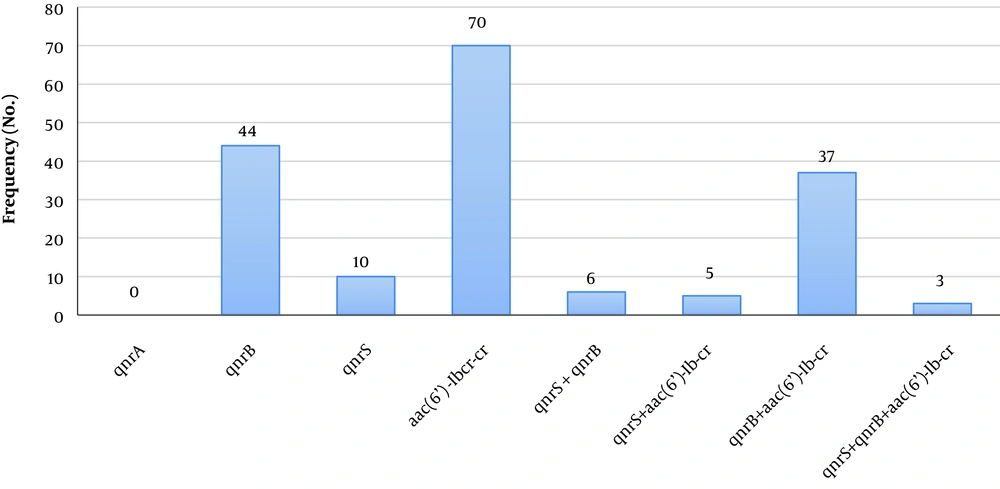
Furthermore, PMQR genes were detected in 62.7% (n = 79) of 126 K. pneumonia isolates, of which 80.76% (n = 42) isolates were ESBL-producing. aac(6’)-Ib-cr was the most common gene resistant against quinolones (n = 70, 55.6%), followed by the qnrB (n = 44, 34.9%). Moreover, the simultaneous presence of resistance genes was observed in 37 strains with the qnrB and aac(6’)-Ib-cr (Figure 1). ESBL-producing samples correspondingly revealed more frequency regarding these genes (Table 3). These results indicated a significant correlation between the qnrB and aac(6’)-Ib-cr genes and resistance against most of the evaluated antibiotics, especially cephalosporins and aminoglycosides (P < 0.05). The PCR results for the detection of these genes are described in Figure 2.
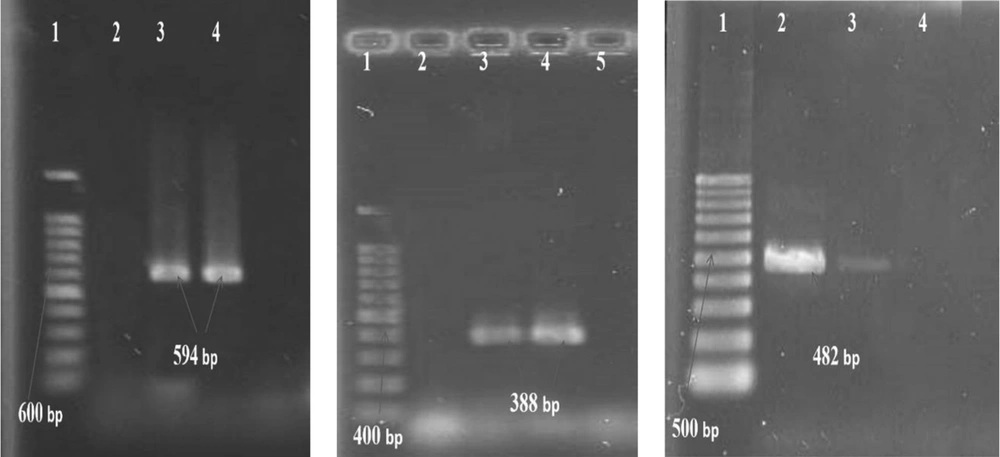
(Gel electrophoresis of PCR products of qnr) qnrB: 1- Lader (100 bp), 2- Negative control, 3- Positive control (594 bp), 4- Positive sample (594 bp): qnrS: 1- Lader (100 bp), 2- Negative control, 3- Positive sample (388 bp), 4- Positive control (388 bp): aac(6’)-Ib-cr: 1- Lader (100 bp), 2- Positive control (482 bp), 3- Positive sample (482 bp), 4- Negative control
5. Discussion
Klebsiella pneumonia is an opportunistic emerging pathogen that causes nosocomial infections. Quinolones are commonly used for treating infections caused by the Enterobacteriaceae species, including K. pneumonia. On the other hand, increased resistance to these antibiotics is associated with severe therapeutic outcomes (16). The present study aimed to evaluate the prevalence of quinolone resistance genes in ESBL-producing K. pneumoniae strains isolated from burn wounds. Most of the K. pneumonia positive isolates were found in males aged 31 to 45 years. Of the 126 K. pneumonia isolates, 69 (54.8%) were MDR and 52 (41.3%) ESBL-producing. According to the literature, the prevalence of ESBL-producing strains in Iran ranges from 12 to 72% (17-19). More than 88% of positive ESBL isolates were MDR.
Based on the previous studies, the frequency of MDR in the ESBL-producing K. pneumonia isolates ranges from 63.33 to 92%. That is consistent with the findings of the current study (5, 20-22). Accordingly, more than 86% of ESBL-producing strains were resistant against cephalosporins. Meanwhile, a study conducted in Iran has reported that almost all ESBL-producing strains were resistant to these antibiotics (23). Based on the findings, in the present study, the resistance of the ESBL-producing strains against aminoglycosides (68.3%) was higher than those reported by Goudarzi et al. (46.35%), Shams et al. (45%), and Eftekhar (36.7%) (10, 13, 22). Moreover, more than 50% of the ESBL-producing K. pneumonia isolates were resistant against fluoroquinolones, and the highest and lowest resistance was, respectively, found against nalidixic acid (65.4%) and gatifloxacin (40.4%). According to studies conducted in Iran, 37.5 - 80% of ESBL-producing K. pneumonia isolates are resistant to ciprofloxacin (10, 22, 24). In the present study, resistance against this antibiotic was 59.6% in ESBL-producing isolates. The global trend of resistance to fluoroquinolones in ESBL-producing bacteria is on the rise (25). Nevertheless, these conflicting results may be due to the difference in antibiotic susceptibility patterns, treatment regimens, types of isolates, geographical differences, and variations in health care control systems at medical centers (3).
PMQR genes play a significant role in resistance against quinolones and fluoroquinolones among ESBL-producing K. pneumonia isolates. The association between PMQR genes with resistance to quinolone and antibiotic-resistant K. pneumonia is reported by several studies (26). Most of the resistant isolates were found in K. pneumonia isolates (27, 28). In the present study, 79 (62.7%) out of 126 K. pneumoniae isolates harbored PMQR genes. However, PMQR determinants were found in 80.76% of ESBL-producing isolates. Based on the studies conducted in Iran, the prevalence of PMQR genes in K. pneumonia isolates ranges from 39.5 to 89.1% (10, 13, 26, 27, 29, 30), while in other countries it ranges from 11.1 to 100% (e.g., South Korea, the United States, and Syria) (31-33). Generally, susceptibility to quinolones and aminoglycosides is lower in species harboring the acetyltransferase aac(6’)-Ib-cr gene. In this study, the most common plasmid gene in K. pneumonia strains was the aac(6’)-Ib-cr (55.6%), as reported by several studies (34).
In the study by Kim et al., performed in South Korea, 55.3% of the isolates (n = 85) were carrying the aac(6’)-Ib-cr gene, which is consistent with the findings of the present study (35). In other studies performed by Goudarzi in Iran (city of Tehran) and Yang in Korea, the prevalence of aac(6’)-Ib-cr gene is reported as 68.8% and 77.5%, respectively, which is higher than the prevalence reported in the present study (13, 31). Moghadam et al. reported a prevalence of 31.8% for aac(6’)-Ib-cr gene (20). It worth noting that PMQR is mediated by the qnr genes, which can promote the rapid development of antibacterial resistance in the Enterobacteriaceae species due to being located on various integrons (9). In the current study, qnrB was the dominant gene (34.9%) of qnr genes. According to the studies performed in Iran, the prevalence of qnrB gene ranges from 1.6% - 88.9% (10, 13, 22). While studies conducted in other countries reported the qnrB as the most common qnr gene in K. pneumonia species (36-38).
In the present study, none of the isolates was carrying the qnrA gene, but in the study by Moghadam et al., the frequency of this gene is determined as 5.6% (20). Similar to the results of the current study, Hassuna et al., in a study performed in Egypt, couldn’t detect qnrA gene in K. pneumoniae isolates (39). The co-presence of the aac(6’)-Ib-cr and the qnrB genes in E. coli and K. pneumonia samples is also reported in various conducted all around the world (27, 40-42). The isolates carrying both genes are more resistant to aminoglycosides, cephalosporins, and quinolones, compared to those only harboring the aac(6’)-Ib-cr gene, which highlights the pivotal role of the qnrB in forming this type of resistance. Other resistance mechanisms, such as mutations in the gyrA and gyrC genes or existing of QepA inoculation pump, may also be responsible for high antibiotics resistance (43). In the present study, 37 (29.3%) isolates were simultaneously harboring qnrB and aac(6’)-Ib-cr genes. In studies performed by Alheib (in Syria) and Eftekhar (in the city of Tehran, Iran), 8 (33.3%) and 21 (50%) isolates were simultaneously carrying aac(6’)-Ib-cr and the qnrB genes, indicating a higher prevalence of antibiotic resistance compared to what was observed in the present study (10, 33).
Based on the study findings, the highest frequency of the qnr genes was observed in quinolone-resistant isolates. The highest prevalence rate of these genes was also reported in isolates resistant to quinolone (37). In the ESBL-producing K. pneumonia strains examined in the present study, the frequency of the qnrB and qnrS genes was 46.1% (24 isolates) and 11.5% (6 isolates), respectively. In ESBL-producing isolates studied by Dehghan Benadkouki et al., 30 were carrying only one of the qnr genes, including the qnrB (n = 12, 45.7%) and qnrS (n = 7, 15.3%), which to a great extent is in line with the findings of the present study (23). According to Shams et al., 51.7% of ESBL-producing isolates were harboring the qnr genes (22). The findings of studies in the United States, Malaysia, and China on the prevalence of qnr genes in ESBL-producing K. pneumonia isolates indicated a prevalence of 11.1, 48.9, and 65.5%, respectively (32, 37, 44).
5.1. Conclusions
This study showed high resistance to aminoglycosides and cephalosporins, as well as a comparatively high prevalence of fluoroquinolone-resistance genes in ESBL-producing K. pneumonia strains collected from burn patients in the city of Kermanshah (Iran). Also, ESBL-producing K. pneumonia isolates showed higher antibiotic resistance and more qnr genes were detected among them. Since burn patients experience severe life-threatening infections and due to the high antibiotic resistance in bacterial isolates inducing such infections, the results of this study are useful for developing plans to address the spread of resistant strains as well as the controlling antibiotic resistance.