1. Background
Pseudomonas aeruginosa is a ubiquitous opportunistic pathogen that is commonly detected in hospitalized patients, especially patients admitted to the intensive care unit (1). Prior exposure to β-lactams, carbapenems, and quinolones has caused the development of multidrug-resistant (MDR) P. aeruginosa, which is associated with high mortality and has led to a worldwide public health problem (2). In particular, the ability of P. aeruginosa to form biofilms enhances its tolerance to antimicrobial therapy (3). It has been reported that P. aeruginosa biofilms grown on implanted and indwelling medical devices could efficiently prevent antimicrobial penetration and protect bacteria from host defense (4). Therefore, developing novel therapies, specifically against MDR P. aeruginosa biofilms, is an urgent need.
A space environment provides a unique set of physical conditions, including microgravity (MG), cosmic radiation, and low nutrients (5). Microorganisms always demonstrate high adaptability to the space environment through phenotypic and molecular alterations (6). There is a general agreement that MG has a major influence on the physiology of bacteria inside a spacecraft during space travel (7). Notably, compared to normal gravity (NG), weightlessness has been shown to decrease the biofilm formation ability of some bacteria, although the specific mechanism remains unknown (8). Since the biofilm formation ability of bacteria is closely related to their antibiotic resistance, weightless technology might serve as a novel tool in the search for therapeutics and preventive strategies against infectious diseases induced by MDR bacteria (9). However, considering that bacterial samples under the MG condition during spaceflight are extremely confined due to safety considerations and logistic reasons, the effects of MG on microbes are usually analyzed with the help of a three-dimensional clinostat, which is an effective instrument to simulate MG on the ground.
The clinostat is designed for decreasing fluid shear and offsetting gravity on the ground. Microorganisms in the equipment rotate on a horizontal axis parallel to the ground and experience gravity that is similar to actual weightlessness. Several studies have shown that microbes exhibited the same characteristics under ground-based MG analogs as they did under the real MG condition. For example, Salmonella typhimurium grown under the MG condition showed an increased virulence in mice. This phenomenon was expected due to a prior ground-based experiment that utilized a rotating bioreactor and produced similar results (10, 11). Therefore, in the present study, we employed a clinostat to simulate MG on the ground, observed MDR P. aeruginosa biofilm formation ability, and explored the relevant mechanism using transcriptomic analysis after two weeks of cultivation.
2. Objectives
This study aimed to investigate the imulated microgravity (SMG) effects on MDR P. aeruginosa biofilm formation and explore the relevant mechanism.
3. Methods
3.1. Acquisition of Strain and Culture Condition
The original strain of P. aeruginosa was obtained from the sputum sample of a 78-year-old male patient with ventilator-associated pneumonia. The grown colonies from the sample were cultured for aerobic bacteria on a blood agar plate and an eosin methylene blue (EMB) agar plate. This clinical isolate was first identified by biochemical tests with the API20NE identification system and subsequently confirmed by 16S rRNA gene sequencing. The antimicrobial susceptibility test of the P. aeruginosa isolate was determined by the disk diffusion method according to the Clinical Laboratory Standard Institute (CLSI 2016) guidelines (12). The results showed that it was resistant to ciprofloxacin, amikacin, piperacillin-tazobactam, gentamicin, cefepime, levofloxacin, imipenem, meropenem, tobramycin, and ceftazidime. The strain was cultivated at 37°C for 24 h in the Luria-Bertani (LB) medium. Then, the cultures were adjusted to a final concentration of 104 CFU/mL and inoculated into 25 cm2 culture flasks. The air bubbles of the flasks were carefully removed. The cultures (referred to as PAM) were placed in a three-dimensional clinostat designed by the China Astronaut Research and Training Center to simulate an MG environment. The cultures under the SMG condition were grown at 37°C, rotating at 30 rpm for two weeks. Except for clinorotation, the NG group (referred to as PAG) was handled in the same way as the SMG group.
3.2. Phenotypic Analysis
3.2.1. Growth Rate Assay
The growth rate assay was performed as described previously (8), with some modifications. Both PAM and PAG were grown to a density of 108 CFU/mL, and 20 µL of a bacterial suspension was inoculated into a 96-well microtiter plate containing 350 µL of fresh liquid LB medium. The growth rate of each sample was monitored by measuring the optical density (OD) at 600 nm every 2 h with the Bioscreen C system (Lab Systems, Finland). A blank well containing 370 µL of LB liquid medium was established as a negative control.
3.2.2. Swimming Motility Assay
The swimming motility assay was performed similarly to the method described previously (13). Each sample was grown to a density of 108 CFU/mL, and 1 µL suspension was used to inoculate the swimming agar (1% tryptone, 0.5% yeast extract, and 0.3% agar). After incubation at 37°C for 24 h, the diameter of the swimming zone was measured for each strain.
3.2.3. Biofilm Formation Assay
The biofilm formation assay was performed as described previously (8), with some modifications. Approximately 200 µL of each sample, with the required concentration of 108 CFU/mL, was inoculated into a 96-well microtiter plate and statically cultivated at 37°C for 24 h. The wells were washed three times with PBS to remove unattached cells. The 96-well microtiter plate was then cultured at 80°C for 15 min to fix the biofilm. The adhered cells were stained with 200 µL of 0.1% crystal violet for 15 min. Subsequently, the 96-well microtiter plate was rinsed with PBS and dried for an additional 10 min. Finally, the stained wells were solubilized in 95% ethanol and the OD570 value for each well was measured by a Thermo Multiskan Ascent (Thermo, USA).
3.3. Transcriptomic Sequencing and Comparison
3.3.1. Sequencing and Filtering
The total RNA was extracted from the samples using the RNeasy Protect Bacteria Mini Kit (QIAGEN, German) according to the manufacturer’s instructions. The quality and quantity of the extracted RNA were measured by the concentration and OD260/280 of samples using Nanodrop One (Thermo, USA). The purified mRNA was then cleaved into short fragments (100 - 500 bp) using a fragment buffer. These fragments acted as templates, and the first strand of cDNA was synthesized by random hexamer-primer. Subsequently, after removing the dNTPs, the second strand of cDNA was generated using RNase H and DNA polymerase I. Short fragments of cDNA were purified with a PCR extraction kit and resolved with EB buffer. After end repair and poly-A addition, the cDNA fragments were connected using sequencing adapters. The UNG enzyme was used to degrade the second-strand cDNA, and the product was purified using a MiniElute PCR Purification Kit before PCR amplification. Finally, the library was sequenced using Illumina HiSeq2000. Raw data were filtered and clean reads were acquired by following three steps: removing reads with adaptors, removing reads with unknown nucleotides ≥ 10%, and removing reads with 40 bp of low-quality bases (≤ Q20).
3.3.2. Gene Expression Value and Differential Expressed Genes (DEGs) Statistics
The gene coverage was estimated by mapping the paired-end reads to the reference reads using SOAP aligner/soap2 software. Reads Per kb per million reads (RPKM) were provided to calculate the gene expression values, and the false discovery rate (FDR) control was provided to obtain the true number of the differential expressed genes (DEGs). The DEGs were defined as those with an RPKM ratio of the two samples of > 2 and an FDR ≤ 0.001. Pathogenicity analysis of DEGs was performed with the virulence factors of pathogenic bacteria (VFDB) database.
3.4. Statistical Analysis
The phenotypic experiments were independently performed in triplicate, and the data were represented as the mean ± standard deviation (SD). A statistical comparison of the data was conducted using the two-tailed Student’s t-test. Statistical significance between the two groups was defined as P ≤ 0.05.
4. Results
4.1. Phenotypic Characteristics
4.1.1. Growth Rate Analysis
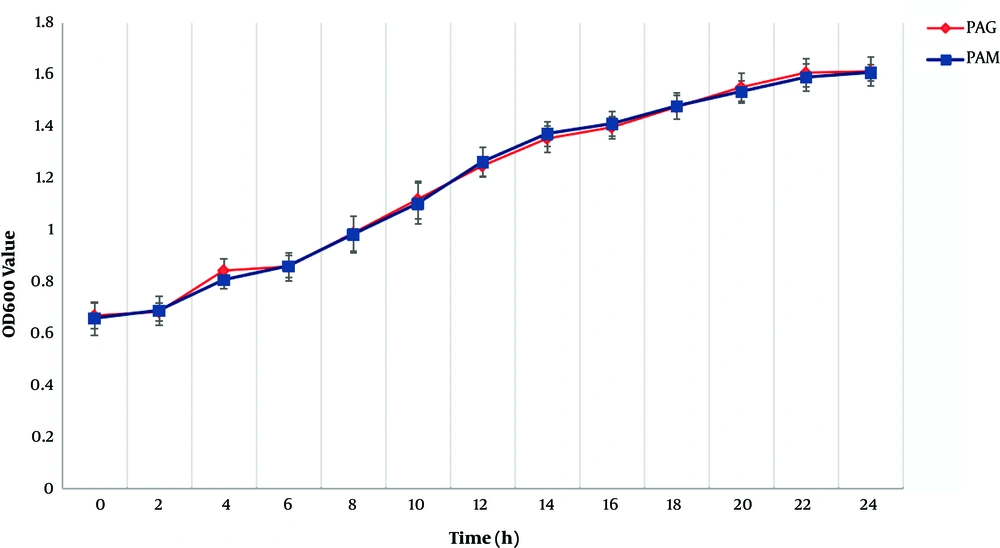
Growth rate data for PAM and PAG are shown in Figure 1. Compared to PAG, PAM experienced a similar growth rate, indicating that the SMG condition did not change the growth rate of P. aeruginosa compared to the NG condition.
4.1.2. Swimming Motility Assay
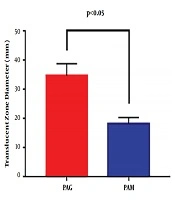
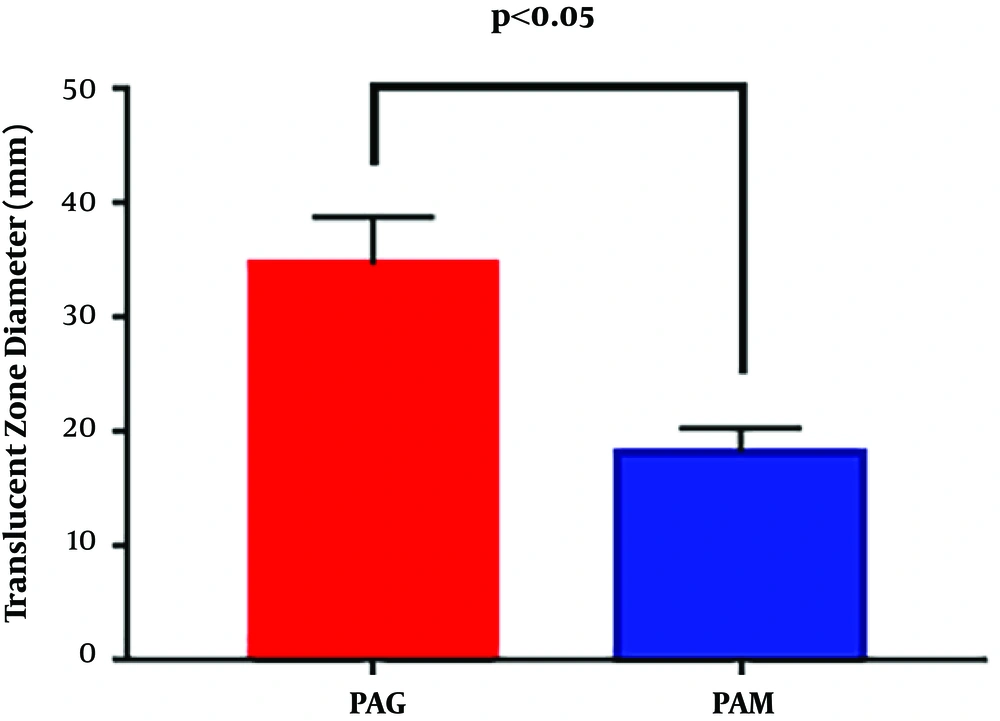
The swimming motility of PAM and PAG was determined by measuring the diameter of the translucent zone formed by the bacteria. The results showed that the swimming motility of PAM was greatly decreased compared to that of PAG (18.33 ± 2.08 mm versus 34.67 ± 4.16 mm, P = 0.0054), suggesting that the SMG condition reduced the swimming motility of P. aeruginosa in contrast to the NG condition (Figure 2).
4.1.3. Biofilm Formation Analysis
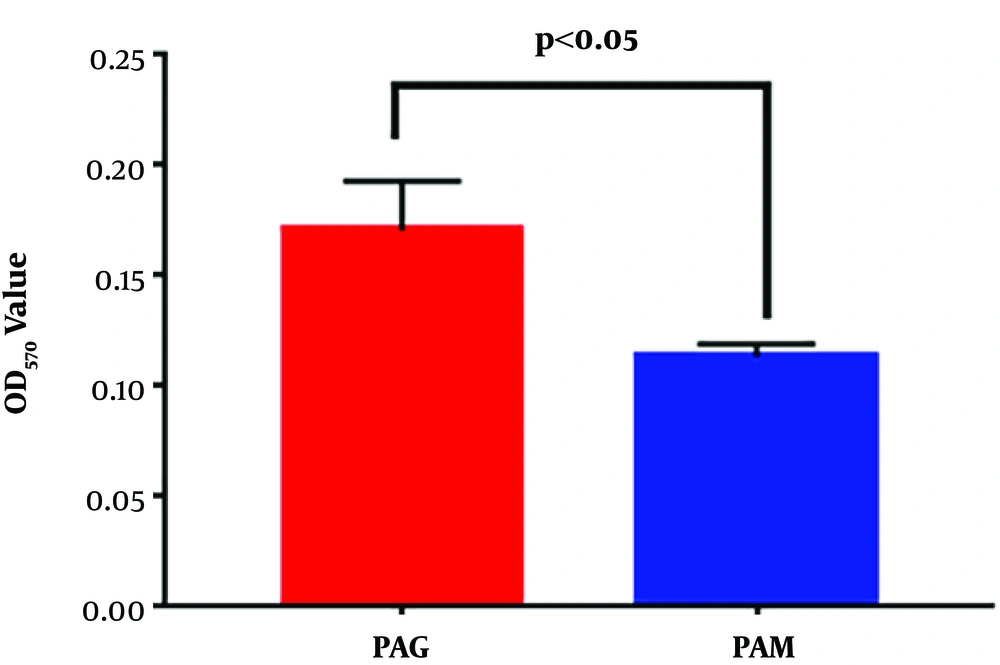
The biofilm formation ability of PAM and PAG was further quantified by monitoring the OD570 values of crystal violet staining. Quantitative results showed that PAM exhibited a decreased OD570 value when compared to PAG (0.171 ± 0.022 mm versus 0.114 ± 0.006 mm, P = 0.0275) (Figure 3). Thus, the results revealed that the SMG condition reduced the biofilm formation ability of P. aeruginosa.
4.2. RNA-Seq Mapping and Comparative Genomic Analysis
Transcriptomic data have been uploaded to GenBank, and the accession numbers of PAM and PAG are SAMN12855527 and SAMN12810912, respectively. A total of 4199 DEGs were identified in PAM, including 330 upregulated genes and 3869 downregulated genes. Compared to PAG, the ratio of downregulated genes to upregulated genes was approximately 11.72, suggesting that gene expression was inhibited in PAM. The expression of flagellar and type IV pili biogenesis genes, fimU, flhB, fliQR, and pilDEXY1Y2VW, were downregulated in PAM (Table 1). The downregulation of these genes might be related to the decreased swimming motility and biofilm formation ability of this P. aeruginosa strain under the SMG condition.
| Gene | Production | Expression |
|---|---|---|
| flhB | Flagellar biosynthetic protein FlhB | Down |
| fliR | Flagellar biosynthetic protein FliR | Down |
| fliQ | Flagellar biosynthetic protein FliQ | Down |
| pilD | Type IV4 prepilin peptidase PilD | Down |
| fimU | Type IV pili biogenesis protein FimU | Down |
| pilV | Type IV pili biogenesis protein PilV | Down |
| pilW | Type IV pili biogenesis protein PilW | Down |
| pilX | Type IV pili biogenesis protein PilX | Down |
| pilY1 | Type IV pilil biogenesis protein PilY1 | Down |
| pilY2 | Type IV pili biogenesis protein PilY2 | Down |
| pilE | Type IV pili biogenesis protein PilE | Down |
| pilD | Type IV prepilin peptidase PilD | Down |
5. Discussion
In recent years, the increasing rates of MDR P. aeruginosa among hospitalized patients have become a major public health problem. Although some biological agents have been used for targeting biofilms induced by P. aeruginosa, such as monoclonal antibodies, the clinical efficacy and safety of these compounds have not yet been sufficiently evaluated (14). Changes in drug-resistance genes and regulatory transcription factors have been observed after microbial exposure to space and SMG environments, which provides a new platform for novel microbiological research that may accelerate the development of new antimicrobial agents leading to drug resistance (5).
In this study, we exposed an MDR P. aeruginosa strain to an SMG environment for two weeks using a clinostat and analyzed its phenotypic characteristics, including the growth rate, swimming motility, and biofilm formation ability. The results indicated that MDR P. aeruginosa cultivated under the SMG condition exhibited decreased swimming motility and biofilm formation ability. Further analysis indicated that the decreased swimming motility and biofilm formation ability might be attributed to the expression of downregulated genes associated with flagella synthesis (flhB, fliQ, and fliR) and type IV pili biogenesis (pilDEXY1Y2VW). This is the first study about MDR P. aeruginosa under an SMG environment, and the identification of targets will provide new directions for drug development and refractory infection treatment strategies.
Swimming motility is a flagella-dependent form of movement observed in the Gram-negative bacterium, P. aeruginosa (15). It promotes the attachment of P. aeruginosa to surfaces and the aggression to other parts of the host, thereby making the site of infection expanded and the degree of infection exacerbated (16). Several studies have demonstrated that the flagellum of P. aeruginosa regulates bacteria motility, allows the bacteria to attach to the surface of the body or materials, and plays an important role in biofilm formation (17, 18). In this study, the swimming motility of PAM was significantly reduced compared to that of PAG, indicating that the SMG condition increased the motility of MDR P. aeruginosa compared to the NG condition. Furthermore, the mechanism behind the decreased motility of MDR P. aeruginosa under the SMG condition was explored by comparative transcriptomic analysis.
Three flagella synthesis-related genes, including flhB, fliQ, and fliR were downregulated in P. aeruginosa cultivated under the SMG condition. The flhB gene played an important role in the determination of the flagellar hook protein and the flagellar filament protein (19). It has been reported that flagella synthesis and motility were completely abolished in the flhB gene deletion mutants (20). The genes fliQ and fliR are likely the components of the flagellar export apparatus and may be physically related to the flagellum itself (21). The fliQ and fliR genes also encode membrane proteins, which play a role in the early stages of flagellar biogenesis and belong to a family of proteins implicated in the export of virulence factors (22). Therefore, we speculated that the decreased motility of MDR P. aeruginosa under the SMG condition could be attributed to the downregulated expressions of flagella synthesis-related genes of flhB, fliQ, and fliR.
Biofilm formation of P. aeruginosa under the MG condition was first investigated in 2001 and the results showed no significant differences in the morphology of the MG-formed biofilm when compared to the biofilm grown under the NG condition (23). However, the results from another study indicated that P. aeruginosa (PA14) exhibited an increase in the amount of biofilm formation and a novel formation architecture during spaceflight (24). In addition, P. aeruginosa (PA01) grown under the SMG condition also exhibited an increase in biofilm formation ability compared to the NG control (25). The biofilm formation ability of P. aeruginosa under SMG and MG conditions was affected by various factors, including the strain type, shear mode, and nutrient availability. In this study, the biofilm formation ability of MDR P. aeruginosa under the SMG condition was tested using crystal violet staining. Compared to the NG group, the SMG group exhibited a decrease in biofilm formation ability. Further analysis was conducted at the transcriptomic level, and the downregulated expression of a series of pil family genes was detected in the SMG group, including pilD, pilE, pilV, pilW, pilX, pilY1, and pilY2.
It has been reported that type IV pili promote surface and host cell adhesion, colonization, and biofilm formation in P. aeruginosa (26). Type IV pili are composed of thousands of copies of small proteins called major pilins along with the low abundance minor pilins encoded by fimU, pilE, pilV, pilW, and pilX in a polycistronic operon (27). The genes pilEVWX play an important role in the synthesis of minor pilins, the functions of which include priming the assembly, counteracting retraction, contributing to adhesion, and regulating the balance between pilus extension and retraction (28). The gene pilY1 encodes the protein located in the membrane and is involved in the fimbrial assembly, while the gene pilY2 encodes a small protein involved in the fimbrial biosynthesis (29). The gene pilD encodes the prepilin peptidase PilD, which regulates PilE, PilV, PilW, and PilX (30). Therefore, the decreased biofilm formation ability of P. aeruginosa under an SMG environment could be attributed to the downregulated gene expressions described above.
5.1. Conclusions
This study provides data regarding the decreased swimming motility and biofilm formation ability of MDR P. aeruginosa after two weeks of exposure to an SMG environment. Further analysis revealed that the decreased swimming motility and biofilm formation ability could be attributed to the downregulated expression of genes responsible for flagellar synthesis (flhB, fliQ, and fliR) and type IV pili biogenesis (pilDEXY1Y2VW). The availability of the SMG condition allowed us to explore potential links between the bacterial phenotype and molecular biology characteristics, thus opening an appropriate and constructive avenue for medical applications in previously unexplored areas. In the future, genes and gene products contributing to this type of resistance may also be a target and more studies may focus on designing new therapeutics and preventatives against bacterial infectious diseases by disrupting genes associated with virulence and antibiotic resistance.