1. Background
The chromosome of Escherichia coli is normally maintained in a negatively supercoiled state by the activity of DNA gyrase and topoisomerase I (1, 2). DNA gyrase is a heterotetramer enzyme that consists of two GyrA and two GyrB subunits (A2B2). Treatment of cells with antibiotics, which target genes encoding DNA gyrase subunits (gyrA and gyrB) leads to DNA relaxation in E. coli (1). These relaxing conditions are known to promote gyrA and gyrB gene expression. DNA gyrase is one of the important targets for antibacterial drug development. DNA gyrase inhibitors are classified into two groups based on their origin, including natural and synthetic drugs (2). Aminocoumarins, simocyclinone D8, and cyclothialidines are examples of natural drugs. Although they have potent activity, their clinical use is limited due to various reasons, including eukaryotic toxicity, poor efficiency against Gram-negative bacteria, poor penetration into bacterial cytoplasmic cell membrane (2).
On the other hand, synthetic inhibitors examples are microcin B17, CcdB, and fluoroquinolones (a group of quinolones). The first example is a weak inhibitor of DNA gyrase as compared with quinolones. The second one needs ATP for its activity (2). While fluoroquinolones are good and widely used drugs against Gram-negative and -positive bacteria. These antibiotics interact with both subunits of DNA gyrase, namely GyrA (N-terminal domain) and GyrB (C-terminal domain), and trap the enzyme attached to DNA break, thereby preventing DNA religation in a ternary complex (quinolone-DNA gyrase-DNA) (2). However, the development of quinolone resistance limits the clinical efficacy of these antibiotics.
Previous studies have shown that quinolone resistance arises from mutations in target genes (gyrA and gyrB), upregulation of efflux pump, AcrAB-TolC and protection of gyrase with the plasmid-encoded protein QnR (3, 4). To eradicate the quinolone-resistant mutants, there is an urgent need to decrease the synthesis of DNA gyrase subunits at transcriptional levels. In the previous study, the ribonuclease P (RNase P)-based external guide sequence (EGS) technique was used to downregulate gyrA expression (5). In order to design new drugs, it is important to find target proteins, which upregulate the expression of gyrA and gyrB at the logarithmic phase of growth at 37°C. There is no information about this matter. It was known that CspA as a transcription activator increases the synthesis of GyrA under cold shock conditions (10°C) through binding at specific sequence upstream of the gyrA gene (6).
2. Objectives
The aims of this work were to select regulatory proteins, which could bind upstream region of gyrA and gyrB genes using online software and to investigate the expression of gyrA and gyrB genes in mutants, lacking these regulatory proteins.
3. Methods
3.1. Prediction of Transcription Factors (TFs)
TFs are regulatory proteins that bind to target DNA, usually located near promoters and regulate transcription from promoters through interaction with RNA polymerase (7). Approximately 500 nucleotides upstream of the translation start site of gyrA and gyrB genes were considered to find TFs. An online software, Virtual Footprint Promoter Analyzer version 3.0 (http://prodoric.tu-bs.de/vfp/vfp_promoter.php) was used to find TF of gyrA, gyrB, and dnaA genes. The TF sequence binding site with the score of match higher than 6 to given sequence (upstream of gyrA, gyrB, and dnaA genes) were selected for further consideration.
3.2. Bacterial Strains and Growth Condition
We used Lauria Bertani (LB; Merck, Darmstadt, Germany) as the liquid medium and LB agar as the solid medium, prepared by addition of agar (Merck, Darmstadt, Germany) to LB broth. Ciprofloxacin was purchased from Sigma-Aldrich (St. Louis, Missouri, USA). Stock concentration was 10 mg/mL. In addition, the parent strain, E. coli BW25113 and knockout mutants JW3525 (BW25113 ΔcspA::Kanr) and JW2701 (BW25113 ΔfhlA::Kanr) were obtained from the Keio collection (8). The minimum inhibitory concentration (MIC) of the strains was almost similar to the MIC of wild type strain (MG1655) for ciprofloxacin (35 ng/mL). Clones with higher resistance to ciprofloxacin were generated from JW3525 and JW2701 mutants after cultivation on LB broth-containing increasing amounts of ciprofloxacin (9). The resistant derivatives, as well as the MG1655 derivatives with their corresponding resistance, are presented in Table 1.
| Strain/Mutant | Genotype | MIC (µg/mL) | Source/Reference |
|---|---|---|---|
| MG1655 | Wild type | 0.035 | A gift from Prof. R G Lloyd |
| W52 | cspA+ gyrA (Ser83→Leu) | 0.3 | (10) |
| C22 | cspA+ gyrA (Ser83→Leu) marOR (20 bp duplication) | 2 | (10) |
| M2 | cspA+ gyrA (Ser83→Leu) marOR (20 bp duplication) acrAB overexpression | 100 | (10) |
| JW3525 | BW25113 ΔcspA::Kanr | 0.05 | Keio collection |
| HA2 | cspA- | 0.3 | This work |
| JW2701 | BW25113 ΔfhlA::Kanr | 0.045 | Keio collection |
| HA1 | fhlA- | 0.3 | This work |
| HA3 | fhlA- | 2 | This work |
3.3. Sample Preparation for Transcriptional Analysis
For each strain, three fresh colonies were inoculated into separate LB broth tubes and incubated overnight at 37ºC with shaking (180 rpm). They were diluted 1:100 in fresh LB broth-containing different concentrations of ciprofloxacin (0.1 × MIC and 0.5 × MIC) and grown to mid-logarithmic phase (OD600 of 0.5 - 0.6), as described previously (10). Then, RNA protect reagent (Qiagen, Hilden, Germany) was added to cultures and incubated for 5 minutes at room temperature. Cultures were centrifuged, and cell pellets were used for RNA extraction.
3.4. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Assay
Total RNA was extracted from all strains mentioned above using an RNeasy Mini kit (QIAGEN, Hilden, Germany), according to the protocol of the manufacturer. To remove DNA, RNA samples were treated with RNase-free DNase (Fermentas, Waltham, MA), according to the manufacture's protocol. DNase treated RNA was repurified using an RNeasy Mini kit (QIAGEN, Hilden, Germany). RNA samples were amplified by PCR to show the absence of DNA. Primers for soxS gene were used in PCR reaction (Table 2). The purity and concentration of samples were determined using the BiochromTM Ultrospec 1100 spectrophotometer (Thermo Fisher Scientific, Ottawa, Ontario, Canada). Suitable RNA samples should have a ratio of A260/A280 more than 1.8. Then, they were used for the synthesis of cDNA. Reverse transcription was performed using the RevertAid Reverse Transcriptase kit (Thermo Fisher Scientific, Ottawa, Ontario, Canada), random hexamer and Purified total RNA (2 µg).
Abbreviations: F, forward; R, reserve.
The cDNAs obtained from reverse transcription were amplified by PCR reaction with specific primers. Then, they were used to determine the level of gene expression by qRT-PCR using Rotor Gene 6,000 thermocycler (Corbet Research, Sydney, Australia) and the SYBR Green kit (TaKaRa, Otsu, Japan). Primers for gyrB, soxS, and gapA were described in previous studies and presented in Table 2 (9, 11). Primers for gyrA were designed by Primer3 V. 0.4.0 software (http://primer3.ut.ee). Thermal cycling conditions were selected based on previous research, except for the annealing temperature, which varied from one gene to another (10). Melting curve analysis (60 - 95°C) was conducted with continuous fluorescence readings. The relative gene expression was determined using the pfaffl method (ratio of target gene expression to gapA expression) (12). Data related to gene expression are presented as the average of duplicate analyses. Significant differences in gene expression were determined by Student’s t-test (two-paired samples, with two-tailed distribution), using SPSS version 16 software (SPSS Inc., Chicago, IL).
4. Results
4.1. Prediction of TFs for gyrA and gyrB Genes
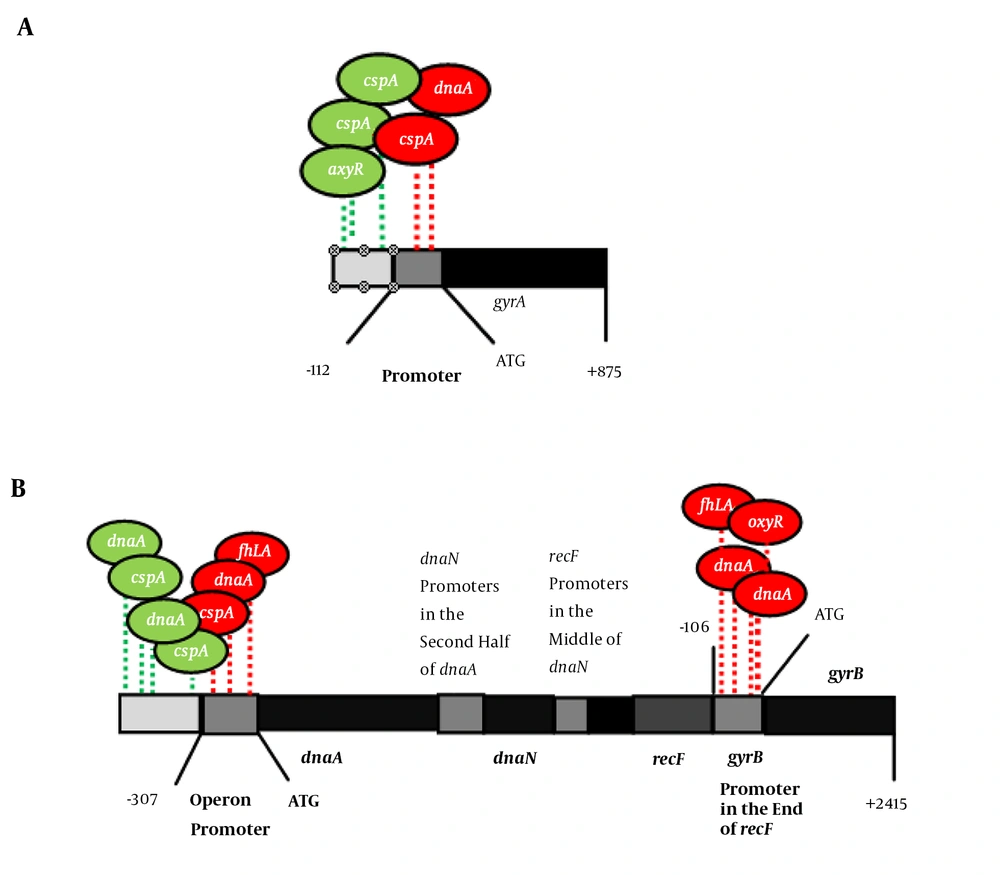
For prediction of TFs for the above genes, online software called Virtual Footprint Promoter Analyzer version 3.0 was used. Sixty-one segments with TF binding characteristics were found for gyrA. Twenty-seven segments were bound to 14 TFs with scores above six (Table 3). Among these TFs, CspA had a score of 10. There were three CCAAT sequences on the negative strand. These sequences are known to be the recognition elements for CspA protein (6) (Figure 1A). It was shown that the overproduction of CspA clearly increases the synthesis of GyrA following cold shock conditions. These conditions also enhance the synthesis of GyrB protein (6). GyrB is located on a four-gene operon (ordered dnaA-dnaN-recF-gyrB). In addition to general promoter, each of these genes has its own promoter(s) (13). There were three CCAAT sequences upstream of above operon (Figure 1B), and CspA might be an activator of this promoter.
| TF | Name | Start | End | Strand | Score | Sequence |
|---|---|---|---|---|---|---|
| ArcA | Aerobic respiration control protein | 348 | 357 | + | 6.4 | TGTTATAATT |
| ArcA | 307 | 316 | + | 6.36 | TGTGAATAAA | |
| ArcA | 131 | 140 | + | 6.16 | GGTTAATGCG | |
| ArgR | DNA arginine binding transcriptional | 303 | 316 | + | 8.86 | TGGATGTGAATAAA |
| ArgR | 346 | 359 | + | 8.61 | TGTGTTATAATTTG | |
| Crp | cAMP receptor protein | 209 | 230 | + | 6.17 | CTTCGTGGTCTACGTTATGGTT |
| Crp | 60 | 81 | - | 6 | AAAGGTGCTCGATGTCGGTTGT | |
| CspA | Cold shock protein | 301 | 305 | - | 10 | CCAAT |
| CspA | 273 | 277 | - | 10 | CCAAT | |
| CspA | 253 | 257 | - | 10 | CCAAT | |
| CytR | Regulator for deo operon | 350 | 361 | - | 7.9 | CGCAAATTATAA |
| CytR | 192 | 203 | - | 7.84 | GCTAAATTTGAA | |
| CytR | 192 | 203 | + | 7.58 | TTCAAATTTAGC | |
| DnaA | Initiation of chromosome replication | 307 | 315 | - | 7.48 | TTATTCACA |
| Fnr | Transcriptional regulation of respiration | 322 | 335 | - | 7.16 | TTGAGGTAAACCTA |
| Fnr | 303 | 316 | + | 6.96 | TGGATGTGAATAAA | |
| IHF | Integration host factor | 280 | 295 | + | 6.21 | AGACAAACGAGTATAT |
| IHF | 141 | 156 | + | 6.14 | GTGCAGCGGTTTGAAC | |
| IHF | 123 | 138 | + | 6.10 | ACGCAGCGGGTTAATG | |
| MetJ | Methionine repressor | 161 | 169 | + | 6.81 | CTTCCAGAT |
| MetR | Regulator for metE and metH | 309 | 315 | + | 9.17 | TGAATAA |
| Mlc | Putative NAGC-like transcriptional regulator | 241 | 246 | - | 6.39 | CGAAAA |
| OmpR | Regulator transcription of ompC and ompF | 239 | 245 | - | 8.7 | GAAAAAT |
| OxyR | Positive regulator of hydrogen peroxide inducible activator | 111 | 156 | + | 13.28 | AATATAGCCCAGACGCAGCGGGTTAATGCGGTGCAGCGGTTTGAAC |
| PdhR | Transcriptional regulator for PDH | 193 | 198 | + | 6.35 | TCAAAT |
| PdhR | 272 | 277 | - | 6.26 | CCAATT | |
| PdhR | 332 | 337 | + | 6.20 | TCAAAC |
The first gene of the operon (dnaA) contained two promoters (14, 15). Based on the results, 83 segments were found for dnaA. Thirty-five segments were bound to 18 TFs, with scores above six (Table 4). One of these TFs is DnaA, which can also bind the upstream region of gyrA and gyrB. For gyrB, there were eight segments bound to seven TFs, with scores higher than 6 (Table 5). FhlA had a score of 10.7. There was also one FhlA binding site in the promoter region of the operon (Figure 1B). FhlA is generally a transcriptional activator of formate metabolism under anaerobic conditions (16). It is claimed that FhlA is not produced in the presence of oxyS mRNA (16). However, the role of FhlA in the expression of gyrA and gyrB genes is not clear. The transcription of oxyS is upregulated due to oxidative stress by OxyR. Both OxyRS and SoxRS are mediated by oxidative stress, induced by ciprofloxacin (17). Based on these theoretical findings, the expression of gyrA and gyrB in the presence and absence of CspA and FhlA, in addition to the expression of the soxS gene in ciprofloxacin-resistant E. coli mutants are described in the following sections.
| TF | Name | Start | End | Strand | Score | Sequence |
|---|---|---|---|---|---|---|
| ArgR | DNA arginine binding transcriptional | 381 | 394 | + | 8.53 | TGGATCTTTATTAG |
| CpxR | 260 | 275 | - | 12.36 | GGAAAAGCGCGGTAAA | |
| Crp | cAMP receptor protein | 291 | 312 | + | 6.82 | GAAAATGTACGACCTCACACCA |
| Crp | 64 | 85 | - | 6.45 | AAGCGCAACCGTTCTCACGGCT | |
| Crp | 130 | 151 | + | 6.37 | AAACTTGAATAAATTCAATGGC | |
| CspA | Cold shock protein | 434 | 438 | + | 10 | CCAAT |
| CspA | 405 | 409 | + | 10 | CCAAT | |
| CspA | 155 | 158 | - | 10 | CCAAT | |
| CytR | Regulator for deo operon | 424 | 435 | - | 7.65 | GGAAAATTTAAT |
| CytR | 254 | 265 | - | 7.54 | GGTAAATAAGGA | |
| CytR | 403 | 414 | + | 7.48 | CAAAAATTGGCT | |
| DnaA | Initiation of chromosome replication | 504 | 512 | - | 7.52 | TTATCCACA |
| DnaA | 270 | 278 | + | 6.44 | TTTTCCGCA | |
| DnaA | 133 | 141 | - | 6.01 | TTATTCAAG | |
| FhlA | Transcriptional activator for fdhf | 648 | 654 | + | 10.14 | TTTTCGA |
| Fnr | Transcriptional regulation of respiration | 134 | 147 | - | 7.40 | TTGAATTTATTCAA |
| Fnr | 134 | 147 | + | 7.26 | TTGAATAAATTCAA | |
| Fnr | 651 | 664 | - | 6.57 | TTGACGTACGTCGA | |
| GcvA | Positive regulator of gcv operon | 390 | 394 | - | 10 | CTAAT |
| GlpR | Glycerol-3-phosphate regulon repressor | 294 | 313 | + | 6.19 | AATGTACGACCTCACACCAG |
| GlpR | 586 | 605 | - | 6.14 | TATGCCCGATCAAGATCCTG | |
| GlpR | 353 | 372 | - | 6.02 | CTTGCGCTTTACCCATCAGC | |
| IHF | Integration host factor | 142 | 157 | + | 6.72 | ATTCAATGGCTTTATT |
| IHF | 88 | 103 | + | 6.02 | GTACAGACGGTTGAA | |
| MalT | Positive regulator of mal regulon | 339 | 344 | + | 8.45 | GGAGGA |
| MalT | 375 | 380 | - | 7.86 | GGACGA | |
| MalT | 716 | 721 | - | 7.82 | GGCGGA | |
| MetJ | Repressor of met genes | 161 | 169 | + | 6.81 | CTTCCAGAT |
| MetR | Regulator for metE and metH | 309 | 315 | + | 9.17 | TGAATAA |
| Mlc | Putative NAGC-like transcriptional regulator | 241 | 246 | - | 6.39 | CGAAAA |
| OmpR | Regulator transcription of ompC and ompF | 239 | 245 | - | 8.7 | GAAAAAT |
| OxyR | Positive regulator of hydrogen peroxide inducible activator | 111 | 156 | + | 13.28 | AATATAGCCCAGACGCAGCGGGTTAATGCGGTGCAGCGGTTTGAAC |
| PdhR | Transcriptional regulator for PDH | 193 | 198 | + | 6.35 | TCAAAT |
| PdhR | 272 | 277 | - | 6.26 | CCAATT | |
| PdhR | 332 | 337 | + | 6.20 | TCAAAC |
| TF | Name | Start | End | Strand | Score | Sequence |
|---|---|---|---|---|---|---|
| Crp | cAMP receptor protein | 9 | 30 | + | 6.5 | CTTTGTCAGCGCGATCAGTGCT |
| CytR | Regulator for deo operon | 54 | 65 | + | 7.78 | CGAAAATTCGAA |
| DnaA | Initiation of chromosome replication | 75 | 83 | - | 6.51 | TTTTCCACG |
| DnaA | 83 | 91 | - | 6.02 | TTTTACCCT | |
| FhlA | Transcriptional activator for fdhf | 53 | 59 | - | 10.7 | TTTTCGT |
| GlpR | Glycerol-3-phosphate regulon repressor | 18 | 37 | - | 6.22 | CGTGTTCAGCACTGATCGCG |
| OmpR | Regulator transcription of ompC and ompF | 55 | 65 | + | 7.92 | GAAAATT |
| OxyR | Positive regulator of hydrogen peroxide inducible activator | 83 | 128 | - | 12.59 | ACGTTTCTCGCTCATTTATACTTGGGTTAATCCGTTATTTTACCCT |
4.2. RNA and cDNA Samples Quality
To ensure that RNA samples were not contaminated with DNA, they were amplified by the PCR method. Figure 2 shows the result of gel electrophoresis of PCR amplification. As can be seen from Figure 2, RNA samples were not contaminated with DNA. The ratio of A260/A280 of RNA samples was more than 1.8. These samples were used for cDNA synthesis. Moreover, cDNAs were used to amplify gyrA, gyrB, and soxS genes. PCR products were electrophoresed on the agarose gel. Figure 3 shows cDNAs were suitable for measuring gene expression by qRT-PCR.
4.3. Expression of gyrA and gyrB in the Presence and Absence of CspA and FhlA
The expression of gyrA gene was measured in low ciprofloxacin-resistant mutants (cspA+ and cspA-) in the presence of ciprofloxacin (0.1 × MIC). The expression of gyrA in a cspA- clone was nearly the same as cspA+ one (P > 0.5) (Table 6). Therefore, the elimination of cspA did not affect the expression of gyrA. The same result was obtained for the expression of gyrB in a cspA- and cspA+ clones (P > 0.5). Inactivation of fhlA did not significantly change the level of gyrB expression in fhlA- clones compared to fhlA+ clones (P > 0.5) in the presence of ciprofloxacin (0.1 × MIC) (Table 6).
| Gene | Wild Type | Mutants | |||||
|---|---|---|---|---|---|---|---|
| W52 | C22 | M2 | HA1 | HA2 | HA3 | ||
| gyrA | 1.0 | 0.8 | 1.3 | 2.6 | - | 1.7 | - |
| gyrB | 1.0 | 1 | 1.2 | 2.5 | 1.5 | 1.5 | 1.6 |
| soxS | 1.0 | 0.8 | 0.9 | 4.8 | |||
aValues represent fold-change (mean of two sets with three samples) in comparison to the wild type strain (MG1655). In all cases, the standard deviation was less than 10% of the mean. Values more than 2 were considered overexpression.
4.4. Expression of gyrA, gyrB, and soxS in the High Ciprofloxacin-Resistant Mutant (M2)
After exposure to high (0.5 × MIC) and low (0.1 × MIC) amounts of ciprofloxacin, the expression of soxS, gyrA, and gyrB increased in the mutant resistant to a high concentration of ciprofloxacin (P < 0.5), but not in mutants showing resistance to low and intermediate concentrations of ciprofloxacin (P < 0.5) (Table 6). This might indicate that at higher concentrations of ciprofloxacin, SoxS could act as a transcriptional activator of gyrA and gyrB.
5. Discussion
One of the essential enzymes for DNA replication is DNA gyrase, which consists of two subunits, GyrA and GyrB. This enzyme is a proper candidate for the development of antibacterial drugs. Among current antibacterial drugs, ciprofloxacin is a suitable antibiotic against Gram-negative bacteria, such as E. coli (2). However, the emergence of ciprofloxacin-resistant E. coli mutants has limited the clinical use of this antibiotic. To overcome this problem, it is necessary to inhibit the expression of antibiotic target genes, encoding two enzyme subunits of gyrA and gyrB. Therefore, the identification of TFs, which upregulate the expression of these genes, is needed to design new drugs against these regulatory proteins. In the present study, it was theoretically proposed that CspA and FhlA might influence the expression of gyrA and gyrB. However, based on experimental findings, they do not play any roles in the regulation of these genes following the treatment with ciprofloxacin.
CspA is a member of cold shock proteins (including CspA, CspB, CspE, CspG, etc.), which are found in Gram-positive and -negative bacteria. They may have overlapping functions, and at least one of these proteins is necessary for bacterial growth. It was shown that cspA transcripts are more abundant in the mid-log phase, while they decrease in the stationary phase to nearly undetectable levels in E. coli in both minimal (M9) and rich (LB) media at 37°C (18). During cold shock, CspA acts as a transcriptional activator both at transcription and translation levels of genes, such as gyrA (18). Our findings, which showed that elimination of cspA did not affect the expression of gyrA and gyrB might imply that regulation of these genes is not controlled by CspA in the presence of ciprofloxacin. It was reported that CspE negatively regulates cspA transcription (19). Therefore, CspE may act as an active cold shock protein in the presence of ciprofloxacin.
Moreover, fhlA gene belongs to hyp operon (formate hydrogenlyase), and its encoded protein, FhlA is involved in formate metabolism as a transcriptional activator under anaerobic condition (20). Our findings, which revealed that inactivation of fhlA did not change the expression of gyrB might indicate that induction of oxidative stress via either OxyRS or SoxRS inactivates FhlA. In addition, low expression of fhlA at a low concentration of ciprofloxacin (3 ng/mL) has been previously reported in a wild type strain (MG1655) (21). Therefore, under aerobic conditions, FhlA does not act as a regulatory protein to enhance the expression of gyrA and gyrB in the presence of ciprofloxacin. Oxidative response is mediated by the OxyR and SoxRS proteins.
SoxRS is closely related to a multiple antibiotic resistance regulator protein (MarA). MarA is upregulated following exposure to ciprofloxacin and activates the AcrAB-TolC pump (22). Ciprofloxacin, in turn, stimulates the production of reactive oxygen species (ROS) in bacterial cells (17). SoxS as a regulatory protein can also enhance the activation of the AcrAB-TolC pump. Our findings that indicating the overexpression of soxS, gyrA and gyrB in the high ciprofloxacin-resistant mutant, indirectly suggest that FhlA may be inactive in the high ciprofloxacin-resistant mutant and that SoxS may play an indirect role in the expression of gyrA and gyrB in the event of oxidative stress.
Furthermore, the finding regarding the overexpression of gyrA and gyrB is consistent with a previous study, which suggested that transcription of topoisomerase genes was sensitive to supercoiling changes and enhanced following the treatment of E. coli cells with quinolone (norfloxacin) and coumarin (novobiocin) (23). However, we could not find any information about the expression of soxS in the mentioned study; this implies that the role of SoxS cannot be excluded. Since there are many DNA binding sites upstream of gyrA and gyrB for regulatory proteins, changes in DNA supercoiling may provide an opportunity for attachment of some regulatory proteins other than CspA and FhlA.
5.1. Conclusions
It can be concluded that under aerobic conditions, the production of ROS, especially by ciprofloxacin, induces SoxS regulon in bacteria, which acts as a strong defense against this antibiotic by activation of AcrAB-TolC efflux pump. It is recommended to use higher concentrations of ciprofloxacin with more caution against Gram-negative bacteria, such as E. coli.