1. Background
Tuberculosis is an airborne disease, primarily caused by Mycobacterium tuberculosis (MTB), affecting vital parts of the body, including the lungs, central nervous system, and lymphatic system (1, 2). According to an estimate, the overall incidence of tuberculosis is reported to be 10 million, accounting for 9.6 - 13.3 million cases in 2018. Moreover, in 2018, the alarming incidence of Multidrug Resistance (MDR) were was 186, 772 cases, and 1.2 million deaths worldwide (3). The risk factors that influence tuberculosis are HIV, diabetes, smoking, alcoholism, and malnutrition (3).
Multidrug-resistant strains mainly emerge due to the development of resistance against the most important and commonly used anti-tuberculosis drugs like rifampicin and isoniazid (4, 5) and contribute mainly to the persistence of relevant infections worldwide (6, 7). Resistance against isoniazid (INH), one of the most effective anti-tuberculosis drugs due to its fastidious potency, is due to genetic aberrations in codons of the hyper-variable region of the katG gene (4). It is also well documented that these mutations usually result in the loss of catalase-peroxidase activity (8, 9), with the majority of them happening in codon 315. However, 30% to 90% of the isoniazid-resistant isolates are associated with the geographical distribution of the population (10).
Isoniazid is one of the drugs inhibiting the biosynthesis of cell wall mycolic acid (long-chain α-branched β-hydroxylated fatty acids) and hence, makes MTB susceptible to oxygen radicals (11). The INH prodrug mostly targets InhA (enoyl-ACP reductase), but it needs activation by the catalase-peroxidase enzyme coded by the katG gene to form reactive free radicals, which then bind covalently to NAD+ to make the isonicotinoyl-NAD complex (12, 13). This complex then binds strongly to the active site of the inhA enzyme, which is actually NADH-dependent enoyl-acyl carrier protein (ACP) reductase and leads to overcrowding of the natural enoyl-AcpM substrate and thus inhibits the biosynthesis of both fatty acids and mycolic acid.
The intrinsic resistance of MTB to hydrophilic antibiotics is believed to be due to the formation of porins in low efficiency in synergy with other resistance mechanisms like enzymatic inactivation or active efflux of drugs. The only peroxide inducible to MTB is KatG, which uses one molecule of hydrogen peroxide (H2O2) as a substrate. However, other molecules of H2O2 act as oxidants and result in the destruction of this toxic substance and lead to the formation of water and oxygen molecules. It has been revealed that catalase activity is essential for the survival of MTB in highly oxidizing environments. The catalytic activity of KatG is mainly due to its structural features, i.e., Met-Tyr-Trp, a covalent cross-link; however, the mutated KatG gene lacks this distinct structural feature (14). Molecular docking can reveal these mutations in the tuberculosis pathogen and may be responsible for drug resistance in isolates from Karachi, Pakistan.
2. Objectives
This study was carried out to analyze the S315R and S315T mutations in MDR-MTB strains of sputum samples concerning their effects on the structure, function, and binding affinity and to determine the extent to which they affect the respective protein. This study is expected to be helpful in the further understanding of the consequences posed by these point mutations and the potential molecular causes of this threatening disease. It will also guide further studies towards the development of better treatment strategies against the havoc posed by different diseases, including tuberculosis. The detailed investigations will also provide valuable insight into some of the features that have not been previously studied.
3. Methods
3.1. Sample Collection
Sputum samples were collected from pulmonary tuberculosis patients at the Ojha Institute of Chest Diseases (OICD), Dow University of Health Sciences, Karachi, Pakistan, after obtaining patients’ consent.
3.2. Staining and Culture
Decontamination of sputum samples was done by using the N-acetyl-L-cysteine sodium hydroxide (NALC-NaOH) concentrated method (15), where NaOH was used as the decontaminating agent and NALC as the mucolytic agent. Sodium citrate attaches to heavy metal ions, which might be present in the samples because these ions attach to NALC and then inactivate it. Sputum and NALC-NaOH samples were then transferred to falcon tubes of up to 5 mL. Next, an equal volume of NALC-NaOH was added to the tubes. Screw caps were tightened, and the tubes were vortexed for 20 s. The tubes were then incubated for 15 min at room temperature and allowed to be decontaminated.
Furthermore, phosphate buffer was added to falcon tubes up to 50 mL and then vortexed, followed by centrifugation for 15 min at 3,000 rpm. The supernatant was poured off by using a funnel into a discordant containing 5% phenol or other mycobacterial disinfectants. A phosphate buffer solution of 0.3 mL was added to sediments to re-suspend them. The sediment of the processed specimen was inoculated into Lowenstein Jenson (LJ) medium using a sterilized pipette, and slides were prepared for Ziehl-Neelsen (ZN) staining. Microscopy was done for the presence of acid-fast bacteria. The media were incubated at 37°C for up to eight weeks or the appearance of any visible colonies, whichever was earlier. After incubation, if cultures yielded no growth, they were declared as negative. Otherwise, they were considered for DNA isolation and further processing.
3.3. DNA Extraction and PCR Amplification
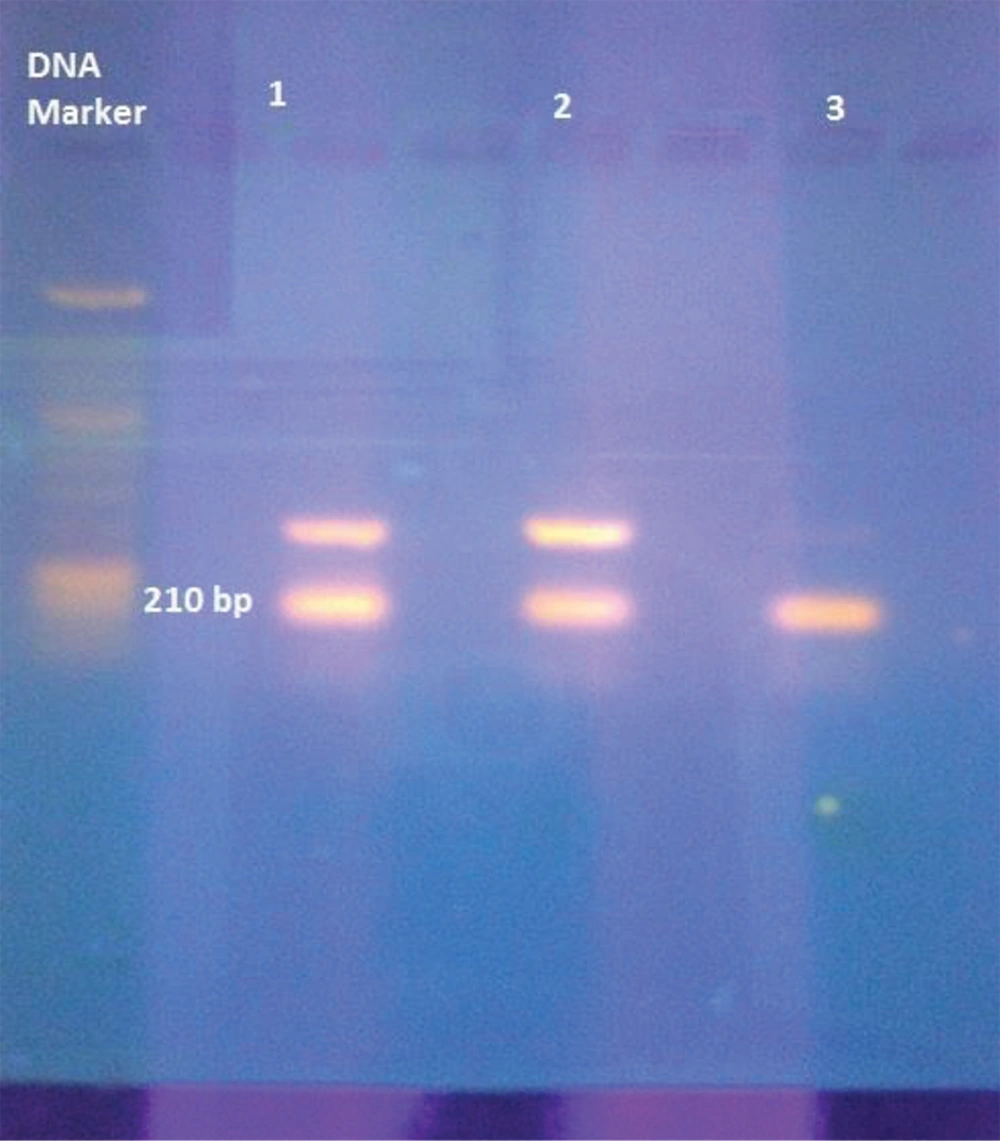
Three to five colonies from solid LJ medium were suspended in 1 mL of molecular biology grade sterile distilled water and were incubated at 95°C for 30 min, followed by 15 min placement in an ultrasonic bath. Tubes containing M. tuberculosis were centrifuged at 13,000 rpm for 10 min at 4°C. The supernatant was taken and was transferred to a new tube. Then, 2 µL of this supernatant was used for PCR analysis (4). The 210 bp segment of the katG gene, a hyper-variable isoniazid resistance region, was amplified using the following synthetic oligonucleotide primers: 5’-GAAACAGCGGCGCTGGATCGT-3’ and 5’-GTTGTCCCATTTCGTCGGGG-3’ (16, 17).
The PCR amplification was done at 94˚C for 10 min for initial denaturation, followed by 40 cycles at 94˚C for one minute, annealing at 45 ˚C for one minute, extension at 72˚C for one minute, and a final extension at 72˚C for seven minutes, in sequence. For visual analysis, gel electrophoresis was performed on the amplified PCR products. To this end, 5 µL of the PCR product was separated through the 2% agarose gel in horizontal gel electrophoresis by supplying an electric voltage of 80 volts. Furthermore, gel staining was done using ethidium bromide (0.5 µg/mL), visualized by UV, and then photographed (3).
3.4. PCR Product Sequencing
As known, PCR-DNA sequencing is a straightforward technology to detect a mutation. For the sequencing purpose, the amplicon was sent to Macrogen Inc., South Korea. The DNA sequencing data were analyzed by comparing with the H37Rv reference strain through the NCBI blast (18-20).
3.5. Datasets
The crystal structure of wild-type MTB with PDB ID:1SJ2 was retrieved from the RCSB Protein Data Bank as a reference. Mutant models of the KatG protein were generated through the mutate model, an application of the modeler available as a python script (21). The optimization of all the predicted models was done using the CHARMM force field while several servers like WHAT IF, ProSA-web, and APOLLO were used for validations (22). The 3D structure of isoniazid was retrieved from PubChem, a database maintained in NCBI (23).
3.6. Docking of Wild-type and Mutated KatG with Isoniazid
Dozens of effective automated docking methods are available for the prediction of biomolecular complexes in structural and functional analyses. AutoDock provides an efficient prediction of bound conformations through a combination of the Lamarckian Genetic Algorithm and an empirical free energy force field. Wild-type and mutant KatG proteins of MTB were, therefore, docked to INH using AutoDock4 (16, 17), a newer version of AutoDock, that integrates the explicit conformational modeling of specified side chains in the receptor and also provides an effective way to analyze the covalently attached ligands. The docked complexes were examined to investigate the potential variations in hydrogen bonding and other interactions. Wild conformations, torsions, and direction of the ligand molecules were set at random. Each docking experiment was derived from 10 different runs that were set to terminate after a maximum of 250,000 energy evaluations. The population size was kept at 150, while a translational step of 0.2 ˚A and quaternion and torsion steps of five were applied throughout the search.
4. Results
4.1. Molecular Analysis
The MDR clinical isolates of patients from Karachi, Pakistan, were found to be resistant to prodrug isoniazid after culturing and susceptibility testing. Strains resistant to isoniazid were screened and further processed to study at a molecular level. The 210 bp covering the hot spot region of the katG gene was amplified via the Nested PCR technique and further examined by gel-electrophoresis (Figure 1).
4.2. Sequence Analysis
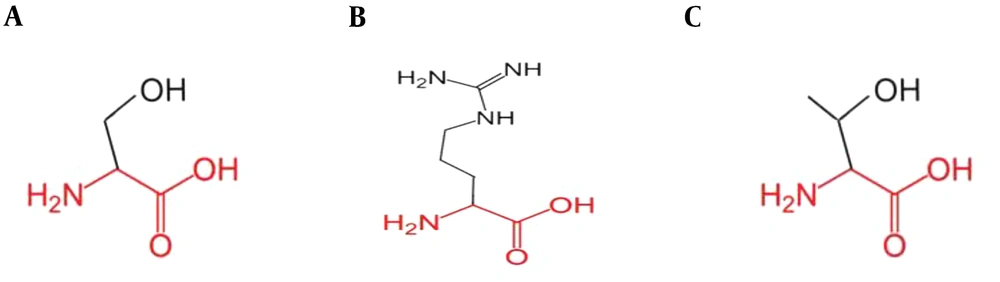
Our investigation revealed mutations to confer isoniazid resistance in clinical isolates, and the examined strains from tuberculosis patients were resistant to the isoniazid prodrug. The 210 bp amplified region of the katG gene covered the hotspot region. The sequencing results of the isoniazid-resistant strains covering codon 315 showed two mutations when comparative BLAST and multiple alignments were run by the online CLUSTAL W2 tool. Two different mutations were observed at codon 315: AGC→ACC (Ser→Thr) and AGC→CGC (Ser→Arg) (Figure 2). Due to these mutations, prodrug isoniazid might not be activated due to improper binding to the catalase-peroxidase enzyme, responsible for drug activation. Our subsequent analysis in isoniazid-resistant isolates was a significant addition to previous studies. This analysis may play an important role in recognizing the isoniazid resistance event. The new alleles found in this study were successfully submitted to DDBJ (NCBI/GenBank) under accession numbers AB776022 and AB776695.
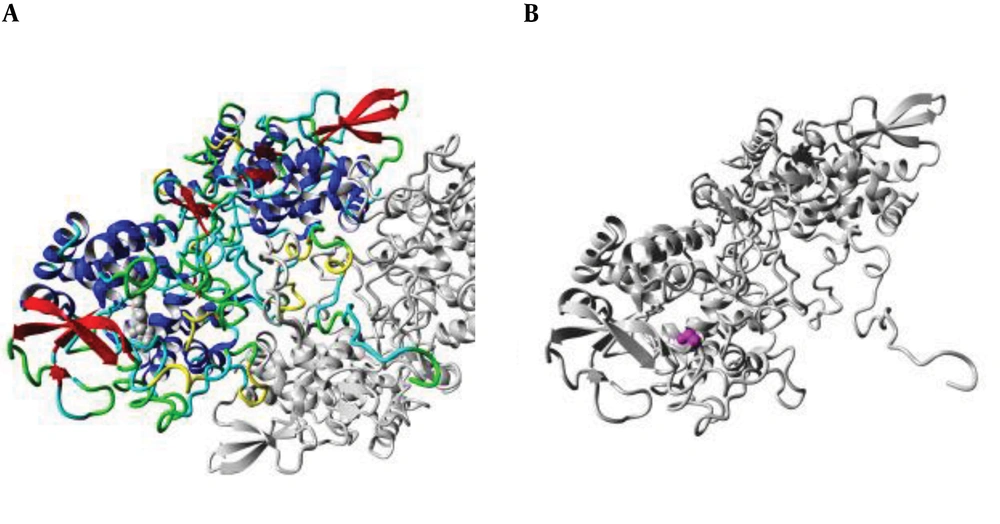
4.3. Structure Analysis
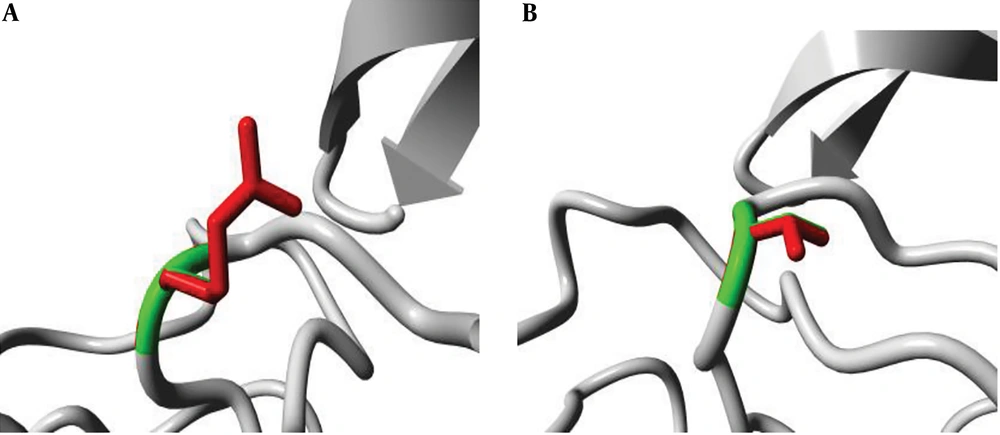
The position of the mutation at codon 315 was analyzed in the ribbon structure of the KatG protein. The protein is colored grey, while the side chain of the mutated residue is colored magenta and shown as small balls. This shows the position of the mutation in the ribbon structure (Figure 3). Our structural analysis further envisages that mutations Serine to Threonine and Serine to Arginine at codon 315 differ in many aspects, especially size and hydrophobic properties. We found that the mutant residue was bigger than the wild one, while mutant residues were comparatively less hydrophobic and bear positive charges, as shown in Figures 4 and 5.
(A) An overview of the KatG protein in ribbon-presentation. The protein is colored by the element; α-helix = blue, β-strand = red, turn = green, 3/10 helix = yellow, and random coil = cyan. (B) The protein is colored grey, and the side chain of the mutated residue is colored magenta and shown as small balls. This shows the position of the mutation in the ribbon structure.
4.4. Docking Studies with Isoniazid Prodrug
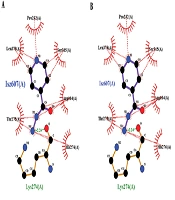
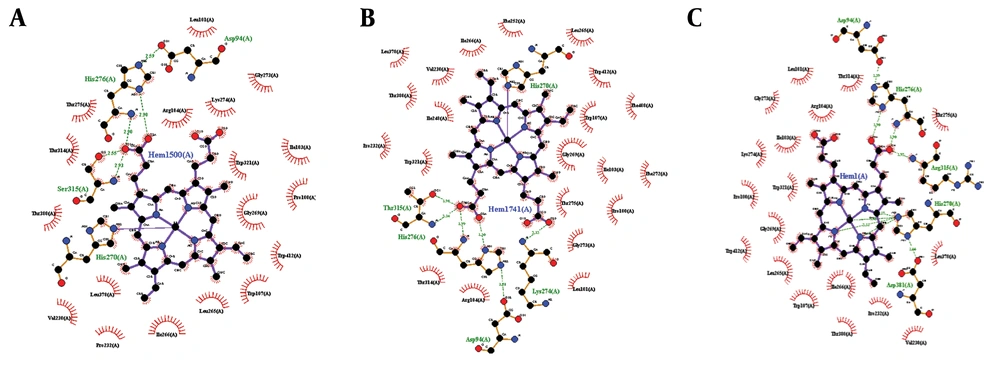
Docking studies were performed through the computational investigation of the docking score and interaction energy between ligand (isoniazid) and MTB KTG protein (1SJ2) retrieved from the protein data bank. AutoDock4 was used for protein-ligand docking, which is an automated molecular docking software package (16). The ligand-protein interaction energies were computed via the PEARLS Program (24), an online resource for energetic analysis of the receptor-ligand system. It computes the total ligand-receptor interaction, electrostatic van der Waals, hydrogen bonds, and solvation free energies along with the ligand-receptor conformational entropy, as shown in Figures 5 and 6. These are physically significant representations of molecules perceived by another molecule in its locality. The negative value of electrostatic energy increased the interaction and vice versa, as shown in Table 1. We observed variations in the bonding patterns of key binding residues of the mutant KatG protein and wild-type KatG (1SJ2). The imprecise bonding pattern of mutated KatG with isoniazid might be responsible for resistance to isoniazid in MTB-tuberculosis strains.
(A) Binding of 1SJ2 with heme; original binding of wild-type 1SJ2 was predicted thorough wet lab. The bonding pattern is as His270 = one covalent link, His276 = two H-bonds, and Ser315 = two H-bonds; (B) Binding of 1SJ2 Ser315Thr with heme. The bonding pattern is as His270 = one covalent link, His276 = two H-bonds, Thr315 = two H-bonds, and Lys274=one H-bond; (C) Binding of 1SJ2 Ser315Arg with heme. The bonding pattern is as His270 = three H-bonds, His276 = two H-bonds, and Arg315 = one H-bond.
| TOS | TLRIE | LRVE | LREE | LRHE | LRSE | LRCE |
|---|---|---|---|---|---|---|
| 1SJ2 wild-type- ISN | -3.80 | -4.17 | -0.81 | -0.23 | 1.32 | 0.09 |
| S315R-ISN | -5.18 | -5.85 | 0.08 | -0.23 | 0.72 | 0.09 |
| S315T-ISN | -4.24 | -5.28 | 0.14 | 0.00 | 0.82 | 0.09 |
Abbreviation: TOS, Type of structure; TLRIE, Total Ligand-Receptor Interaction Energy; LRVE, Ligand-Receptor Van der Waals Energy; LREE, Ligand-Receptor Electrostatic Energy; LRHE, Ligand-Receptor Hydrogen Bond Energy; LRSE, Ligand-Receptor Solvation Free Energy; LRCE Ligand-Receptor Conformational Entropy.
a Energies are in Kcal/mol.
5. Discussion
Isoniazid is a prodrug used as a first-line tuberculosis drug. Different studies have described different mechanisms for isoniazid resistance and have reported multiple mutations in different genes. Different studies have reported that about 50 to 84% of resistance against isoniazid is due to one mutation in ktG whereas 10 - 35% of resistance is due to at least one mutation in the inhA promoter, and 10 to 40% of resistance is due to at least one mutation in oxyR-ahpC (Li et al., 2015). Mutations at katG lead to a high level of isoniazid resistance (25). In our wet-lab study, clinical isolates from the Ojha Institute of Chest Disease, Karachi, Pakistan, were examined, and their susceptibility patterns were investigated. We revealed two important mutations, namely S315T and S315R, in KatG (Figure 2).
A comprehensive investigation about the molecular modeling and dynamics of isoniazid binding with KatG would be helpful for the improvement of therapeutics and can be used against other complex disorders. This study was systematically designed to explore the three-dimensional spatial arrangements and conformational changes at a significant residue contributing to INH binding due to arginine and threonine substitutions. It was observed that the wild-type residue (Serine 315) had interactions with a ligand annotated as heme but significant dissimilarities in the properties of mutant KatG enzyme residues resulted in the loss of the binding affinity to the ligand.
The wild-type residue (Serine) forms a hydrogen bond with isoleucine at position 317, which can be disrupted by the change in the size of wild and mutant-type residues due to the positional changes introduced by mutant residues to make the same hydrogen bond. The mutation is located in a domain primarily important for protein activity and contact with another domain. These interactions could also be disturbed by mutations, which may negatively affect the protein functions including signal transduction between the domains.
There are also differences in charges between the wild and mutant-type residues. The mutants introduce a charge in a buried residue, which is at the core of the protein or protein-complex and potentially would lead to protein folding problems (26). Heme acts as a co-factor in the activation of INH. It was detected that wild-type 1SJ2 Ser315 forms two hydrogen bonds with heme having bond lengths of 2.55 Å and 2.93 Å, and one covalent bond between His270 and Fe, due to mutation S315R, it makes only a single hydrogen bond with the heme molecule with a bond length of 2.93 Å and a covalent bond with His270 and Fe. Variations in the hydrogen bond lengths of 2.91 Å and 3.24 Å were observed in the S315T mutation as compared to the wild type (Figure 5), which may cause inactivation of INH.
Protein-ligand interactions are always of prime importance in biological processes and provide an opportunity to enhance our understanding of protein function and therapeutic interventions (27). Docking calculation described the extent of the interaction of the ligand with the KatG enzyme and the behavior of the ligand-protein complex, as well. In the wild type (1SJ2), the ligand-receptor van der Waals energy was found to be -4.17 kcal/mol. The total ligand-receptor interaction energies of the wild type, mutant S315T, and S315R M. tuberculosis KatG-INH complex were -3.80 kcal/mol, -4.24 kcal/mol, and -5.18 kcal/mol, respectively, as shown in Table 1. The low energy of the mutant complex showed a strong binding affinity between mutant M. tuberculosis KatG and INH. Because of this tight binding, INH will lose the ability to convert itself into an active form (IN-NAD), which is significant for lethal action and staying unsusceptible to mutant KatG.
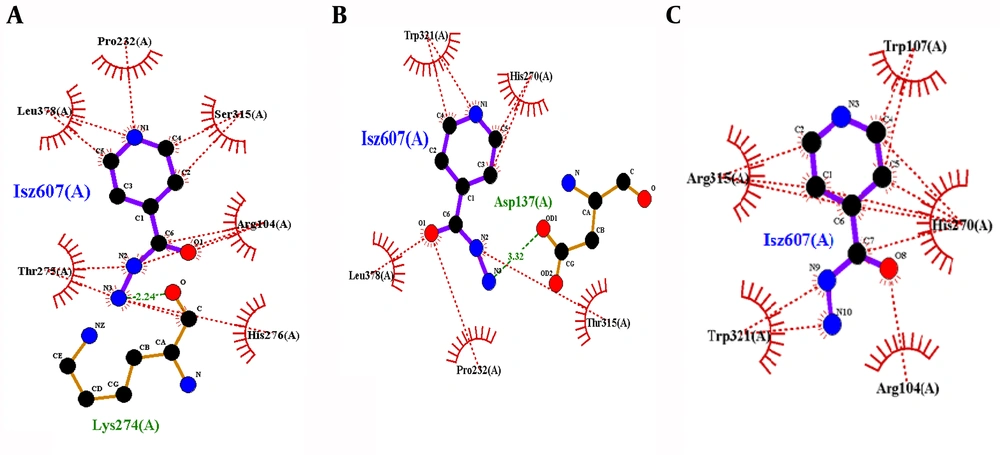
The bindings of mutant KatG residues were observed dissimilar to the wild type, as shown in Figure 7. There was only one hydrogen bond at Lys274 in the wild type and six hydrophobic interactions with residues Arg104, Pro232, Thr275, His276, Leu378, and Ser315. There was no hydrogen bond in mutant S315R KatG, having five hydrophobic interactions at positions Arg104, Trp107, His270, Trp321, and Arg315. Hydrogen bonds were observed distorted in mutant 315, and it is the only similar position between wild and mutant complexes. Furthermore, in mutant S315T, it was observed that a new residue of Asp137 interacted to INH through the hydrogen bond while two new residues of Trp321 and His270 were observed to participate in hydrophobic interactions.
Thus, His270, His276, and Ser315 were seemed to be key binding residues of this binding pocket and were observed to be involved in the drug-protein complex, as shown in Figure 6. Attractions among binding residues and structural arrangements were affected due to the difference in the size and hydrophobic properties of mutant amino acids. The most significant elements responsible for inactivation are changes in binding residues, as observed in the computational model. Indeed, the resistance mechanism against different drugs, including isoniazid is poorly understood. It has been found that different approaches and techniques are used to determine resistance mechanisms associated with mutations in MDR- tuberculosis (24).
This computational investigation showed that variations in binding residues and differences in docking energies due to mutations might be responsible for the inactivation of INH, leading to resistance in M. tuberculosis clinical isolates. Changes in the binding pattern and interacting residues would lose the ability of INH to convert itself into an active form (IN-NAD), which is significant for lethal action and staying unsusceptible to mutant KatG. It was concluded that isoniazid performs better when Ser is at position 315, and the mutation of Arg or Thr at the mentioned position significantly harms the enzyme function. This study will help researchers to better understand the mechanism of resistance development by the bacterium and confirms some of the previously reported results by other researchers. We hope that this data analysis will provide important knowledge relevant to drug resistance and the level of resistance in different MTB strains.