1. Background
Burn injuries continue to be an important cause of disability and mortality in all ages worldwide. Therefore, burns are a global major public health issue. Every year half a million Americans sustain burn infections, that 40,000 of them require hospitalization, and more than 7% of them die. According to the published data, infection with various nosocomial bacteria in burn patients is the sixth leading cause of death in Iran (1, 2). Burn patients, due to their features, are susceptible to colonization and growth of bacteria, which intensify the clinical manifestation and subsequently increases the mortality and morbidity rates (3).
Although various pathogens cause infection in burn patients, Staphylococcus aureus strains are the most prevalent human pathogen that causes infection in these patients worldwide (4-6). Meanwhile, according to the recent data, the prevalence of infection with S. aureus in burn units is increasing every day in both developing and developed countries, which resulted in increased attention to infection control programs in burn patients, particularly in the first 5 days after hospitalization (1). The emergence and ongoing spread of simultaneous resistance to multiple antimicrobial agents among S. aureus strains in burn units is on the rise in Iran, which in turn has increased the treatment failure rates among burn patients with infections and increased the complicacy of treating these patients (7-10).
In general, as a barrier to successfully treat the patients, simultaneous resistance to multiple antimicrobial agents increases challenges related to the treatment of S. aureus infections in burn patients (11, 12). According to the literature, biofilm formation plays a key role in the pathogenesis and drug resistance of S. aureus strains and makes it a highly pathogenic bacterium (11, 13). The biofilm formation of S. aureus is associated with the expression of various microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), including biofilm-associated protein (Bap), fibronectin-binding proteins (Fnb), polysaccharide intercellular adhesion (PIA), and clumping factors (Clf) (14).
2. Objectives
Although several epidemiological studies have investigated the prevalence of S. aureus among burn patients in Iran, phenotypic and genetic characteristics associated with biofilms of these strains are not studied yet. Therefore, gaining adequate knowledge through investigating the prevalence, resistance rates, and the biofilm formation ability of S. aureus strains isolated from burn patients would provide valuable information. The current study aimed to determine the frequency of biofilm-producing genes and antibiotic resistance patterns of S. aureus isolates collected from hospitalized patients in Motahari Burn Hospital, as the main specialized burn hospital in Tehran, Iran.
3. Methods
3.1. Study Design, Identification of Staphylococcus aureus
In this cross-sectional study 83 S. aureus strains isolated from different wound samples of burn patients referred to Motahari Burn Hospital, affiliated to Iran University of Medical Sciences, from December 2017 to August 2018 were investigated. Samples were recovered from the wound areas with the highest degree of burn using sterile swab sticks and scalpel blades. The wound samples were sent to the research laboratory of Shahid Beheshti University of Medical Sciences in the shortest possible time; then samples were cultured on blood agar (BA; Merck; Germany) and incubated at 37°C for 24 h. The standard microbiology method was used to identify phenotypic of S. aureus strains, including Gram staining, culture in mannitol salt agar medium (MSA; Merck; Germany), DNase, coagulase, and catalase test. Definitive identification at species level was performed by polymerase chain reaction (PCR) of targeted S. aureus species-specific fem and nuc genes (15). Confirmed S. aureus isolates were stored in tryptic soy broth (TSB, Merck Co., Germany) containing 20% glycerol at -70°C and were subjected to further molecular analysis.
3.2. Phenotypic and Genotypic Methicillin-Resistant Staphylococcus aureus Screening
All S. aureus isolates were screened for methicillin resistance using the cefoxitin (CEF 30 µg) disc diffusion test based on the Clinical and Laboratory Standards Institute (CLSI 2018) guidelines (https://clsi.org/). Besides, the PCR method was applied for the genotypic amplification of mecA genes. The sequences of degenerated primers used in the current study and amplicon size are listed in Table 1.
| Gene | Primer | Sequence | PCR Condition (Temperature, Time) | Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Initial Denaturation | Denaturation | Annealing | Extension | Final Extension | Number of Cycles | Amplicon Size, bp | ||||
| fnbA | Forward | CACAACCAGCAAATATAG | 95°C, 5 minutes | 94°C, 45 seconds | 55°C, 60 seconds | 72°C, 50 seconds | 72°C, 4 minutes | 35 | 1362 | (16) |
| Reverse | CTGTGTGGTAATCAATGTC | |||||||||
| fnbB | Forward | GGAGAAGGAATTAAGGCG | 95°C, 5 minutes | 95°C, 60 seconds | 55°C, 50 seconds | 72°C, 60 seconds | 72°C, 5 minutes | 30 | 813 | (17) |
| Reverse | GCCGTCGCCTTGAGCGT | |||||||||
| clfA | Forward | GTAGGTACGTTAATCGGTT | 95°C, 5 minutes | 95°C, 45 seconds | 56°C, 60 seconds | 72°C, 60 seconds | 72°C, 3 minutes | 35 | 1584 | (16) |
| Reverse | CTCATCAGGTTGTTCAGG | |||||||||
| clfB | Forward | TGCAAGATCAAACTGTTCCT | 95°C, 5 minutes | 94°C, 45 seconds | 57°C, 45 seconds | 72°C, 55 seconds | 72°C, 5 minutes | 35 | 596 | (16) |
| Reverse | TCGGTCTGTAAATAAAGGTA | |||||||||
| bbp | Forward | CAGTAAATGTGTCAAAAGA | 95°C, 5 minutes | 95°C, 60 seconds | 55°C, 45 seconds | 72°C, 60 seconds | 72°C, 4 minutes | 30 | 1055 | (16) |
| Reverse | TACACGGTGTTGAAGTG | |||||||||
| cna | Forward | AGTGGTTACTAATACTG | 95°C, 5 minutes | 95°C, 45 seconds | 56°C, 60 seconds | 72°C, 60 seconds | 72°C, 5 minutes | 35 | 744 | (16) |
| Reverse | CAGGATAGATTGGTTTA | |||||||||
| ebps | Forward | CAATCGATAGACACAAATTC | 95°C, 5 minutes | 94°C, 60 seconds | 58°C, 45 seconds | 72°C, 60 seconds | 72°C, 5 minutes | 30 | 526 | (16) |
| Reverse | CAGTTACATCATCATGTTTA | |||||||||
| mecA | Forward | AGAAGATGGTATGTGGAAGTTAG | 95°C, 5 minutes | 94°C, 45 seconds | 56°C, 50 seconds | 72°C, 60 seconds | 72°C, 4 minutes | 35 | 583 | (15) |
| Reverse | ATGTATGTGCGATTGTATTGC | |||||||||
3.3. Antibiotic Susceptibility Testing
To evaluate the susceptibility of the isolates (Himedia, Mumbai, India) against erythromycin (ERY), clindamycin (CLI), cephalexin (CEP), and ciprofloxacin (CIP), the Kirby-Bauer disk diffusion method was applied (CLSI 2018). For this purpose, bacterial colonies were dissolved in 2 mL of sterile physiological saline to obtain opacity equivalent to a 0.5 McFarland standard, which is equivalent to a bacterial suspension containing between 1.5 × 108 CFU/mL. Then, the bacterial suspension was cultured on Mueller-Hinton agar (MHA; Merck, Germany). Afterward, antibiotic discs with standard distance were placed on the MHA and incubated for 24 h at 37°C. After incubation, the susceptibility to different antibiotics was conducted based on the CLSI guidelines. Results for cephalexin and ciprofloxacin were interpreted according to the European Committee for Antimicrobial Susceptibility testing (EUCAST) breakpoints (http://www.eucast.org). The resistance of S. aureus strains to three (or more) antimicrobial categories other than beta-lactams was defined as multidrug resistance (MDR) (15). In every test performance, the S. aureus ATCC 25923, ATCC 43300, and ATCC 29213 reference strains were used for quality control.
3.4. DNA Isolation, in Vitro Screening for Biofilm-Related Genes
DNA extraction was performed using the phenol-chloroform method. The quality of extracted DNA was checked on agarose gel for all isolates. The DNA quality was adjusted to approximately 100 ng/µL and calculating the A260/A280 ratio, which was evaluated by a NanoDrop-2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). According to the literature, an A260/A230 absorbance ratio between 1.8 and 2.0 is the accepted range of DNA purity for the performance of the PCR technique (15). All samples were screened for clfA, clfB, fnbA, fnbB, bbp, ebp, and cna genes by PCR assay with oligonucleotide primers as previously described. The oligonucleotide primers and PCR protocols used to determine the presence of the adhesions and biofilm encoding genes are listed in Table 1. For all examined genes, a final volume of 25 µL PCR mixture containing 0.7 μL of each forward and reverse primers (10 pmol), 12 µL of master mix (Cinnagen Co., Iran), including 200 µM for each dNTP, 0.5 M Tris-HCl, 1.5 mM MgCl2, and 0.04 Units/µL Taq, and 10.6 µL of sterile distilled water was used. Finally, a volume of 1 µL of DNA template (100 ng/µL) was added to each 0.2 ml reaction tube.
3.5. Statistical Analysis
Data were analyzed using SPSS version 25.0 (SPSS Inc., Chicago, IL). The Chi-square and Fisher exact tests were used to analyze the data. A P value of < 0.05 was considered statistically significant.
4. Results
A total of 83 S. aureus strains obtained from burn patients hospitalized at a referral specialized burn hospital in Tehran were investigated in this study. Which, 51 (61.4%) patients were male and 32 (38.6%) were female. The median age of patients was 41.6 years, ranging from 17 to 59 years. Age distribution of patients is described in the following: 15 patients were 18 years old or younger, 60 were 18 - 45 years, and 8 were ≥ 60 years old. Based on the results of the cefoxitin disk diffusion method, 83 S. aureus isolates, 73 Methicillin-Resistant S. aureus (MRSA) (87.9%), and 10 methicillin-sensitive S. aureus (MSSA) (12.1%) isolates were identified. the percentage resistant strains was as follow: CEP (87.9%), 75.9% for CLI, 72.3% for ERY, and 60.2% for CIP (Table 2). In total, five resistance patterns were identified, wherein CEF, CEP, CIP, CLI, ERY (36.1%, 30/83), CEF, CEP, ERY (24.1%, 20/83), and CEF, CEP, CLI (15.6%, 13/83) were the top three most frequently identified patterns. Table 3 gives information about the resistance pattern of S. aureus strains collected from burn patients.
| Antibiotic | Staphylococcus aureus | Total | |
|---|---|---|---|
| MRSA | MSSA | ||
| Cephalexin | 48 (87.3) | 0 (0) | 73 (87.9) |
| Cefoxitin | 47 (87) | 0 (0) | 73 (87.9) |
| Clindamycin | 53 (84.1) | 10 (15.9) | 63 (75.9) |
| Erythromycin | 50 (83.3) | 10 (16.7) | 60 (72.3) |
| Ciprofloxacin | 40 (80) | 10 (20) | 50 (60.2) |
| Total | 73 (87.9) | 10 (12.1) | 83 (100) |
Abbreviations: MRSA, Methicillin-resistant Staphylococcus aureus; MSSA, Methicillin-sensitive S. aureus
aValues are expressed as No. (%).
| Simultaneous Resistance to Antibiotics | Resistance Pattern | Number of Isolates, % |
|---|---|---|
| Five | CEF, CEP, CIP, CLI, ERY | 30 (36.1) |
| Four | CEF, CEP, CIP, CLI | 10 (12.1) |
| Three | CEF, CEP, CIP | 13 (15.6) |
| CEF, CEP, ERY | 20 (24.1) | |
| CIP, CLI, ERY | 10 (12.1) |
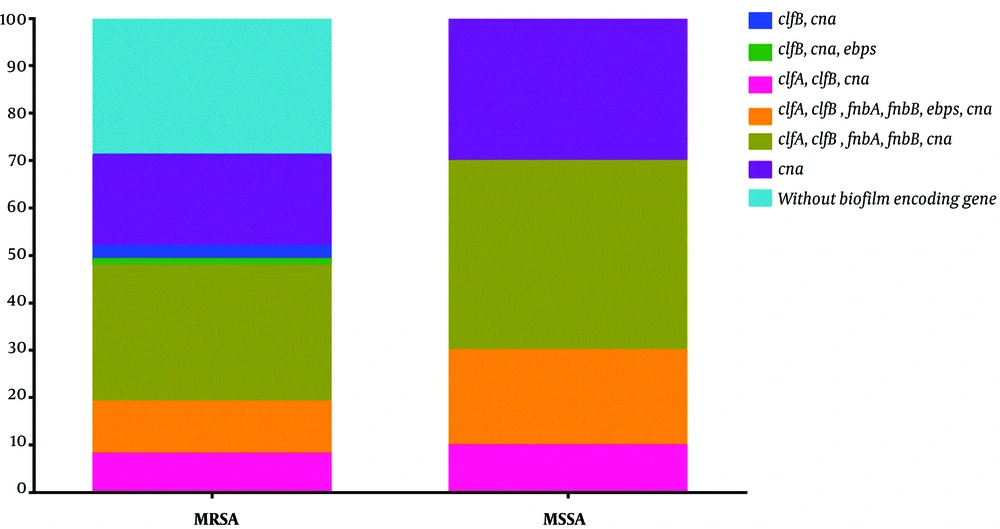
The findings of the genetic evaluation of biofilm and adhesion genes revealed that can, as the most prevalent gene, was present in 62 strains (74.7%), clfB in 45 (54.2%), clfA in 42 (50.6%), fnbA in 35 (42.1%), ebp in 11 (13.2%), and fnbB in 10 (12%) isolates, respectively (Figure 1). None of the MRSA and MSSA strains carried the bap gene. Six different biofilm genetic patterns were identified, wherein clfA, clfB, fnbA, ebp, and cna (30.1%, 25/83), clfA, clfB, fnbA, fnbB, ebp, and cna (12%, 10/83), and clfA, clfB, and cna (8.4%, 7/83) were the top three most frequently identified patterns. The frequency of biofilm encoding genes was higher in MSSA as compared to MRSA strains (100% vs. 71.2%). Data on the distribution of adhesion and biofilm genes among MRSA and MSSA isolates are provided in Figure 2. A significant association was found between the presence of clfA and resistance to erythromycin (P < 0.016) and clindamycin (P < 0.001), clfB and resistance to clindamycin (P < 0.021), fnbA and resistance to clindamycin (P < 0.003), and ciprofloxacin (P < 0.003), cna and resistance to cephalexin (P < 0.001). The chi-square and Fisher exact tests also showed a significant difference between resistance to clindamycin, erythromycin, and cephalexin with the mecA gene (P < 0.05). Moreover, no significant difference was observed among the other examined genes.
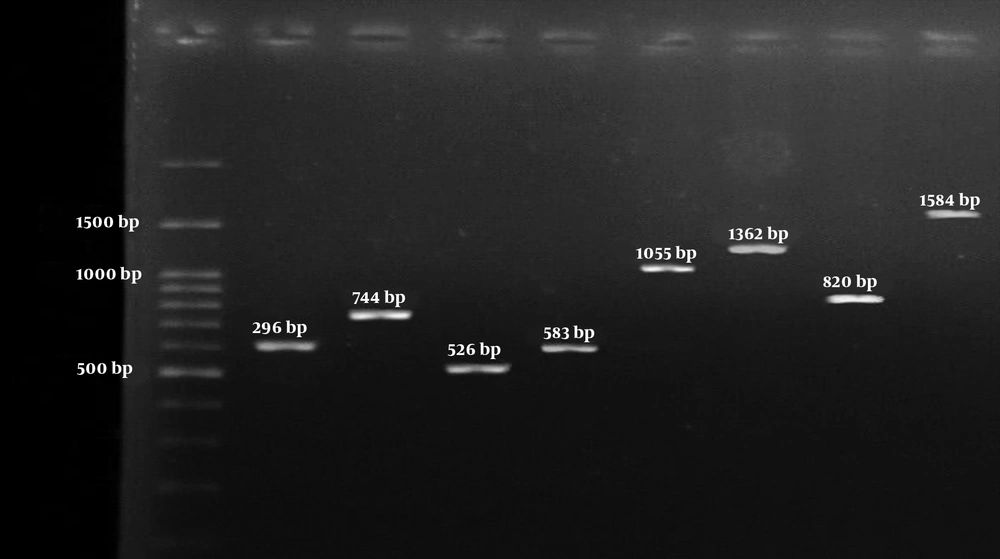
Left to right respectively: 100-bp DNA Ladder (Fermentas, UK); The 596 bp PCR product of clfB encoding gene; The 744 bp PCR product of cna encoding gene; The 526 bp PCR product of ebp encoding gene; The 583 bp PCR product of mecA encoding gene; The 1055 bp PCR product of bbp encoding gene; The 1362 bp PCR product of fnbA encoding gene; The 813 bp PCR product of fnbB encoding gene; and the 1584 bp PCR product of clfA encoding gene.
5. Discussion
Based on several reports, infection with S. aureus is on the rise among burn patients with severe wounds hospitalized at specialized burn hospitals. Therefore, considerable attention has been paid to this issue in recent years (9, 11, 18). Continuous surveillance of infections caused by S. aureus in hospitalized patients not only helps to effectively prevent and control such infections, but it is also important for providing appropriate and effective treatment (1-3). This study showed the high prevalence of biofilm encoding genes among burn patients hospitalized in Iran. Since biofilm formation increases the tolerance to antimicrobial agents and antibiotic resistance, this is a novel and important finding.
There are several factors that affect the frequency of MRSA strains, particularly among patients and in different geographic areas (19, 20). Methicillin-resistant S. aureus screening revealed a prevalence of 87.9%. Studies performed in various countries have reported different prevalence rates for MRSA isolates in burn patients, including Turkey (30.9%), Ireland (54.7%), Israel (46%), Italy (38.3%), Greece (36.6%), France (31.5%), Poland (27.2%), Germany (17.2%), Switzerland (15.7%), and Sweden (2.1%) (4, 21).
According to the findings, the frequency of the mecA gene was 87.9%, which is in line with the studies conducted by Montazeri et al. (88.6%) (22), Nourbakhsh et al. (93%) (23), Hoveizavi et al. (87.36%) (8), and Abbasi-Montazeri et al. (80%) (7). It’s not unexpected that various studies conducted in different countries have reported different rates of prevalence for mecA among S. aureus strains, including in Iran (63.6%), Tunisia (60%), and Nigeria (42.3%) (9, 13, 24, 25). This difference can be attributed to various factors such as study design, study population, and policies about consumption and prescription of beta-lactam antibiotics.
The results also indicated high prevalence rates of resistance to erythromycin (72.3%), clindamycin (75.9%), ciprofloxacin (60.2%), cephalexin (87.9%), and cefoxitin (87.9%). A recent cross-sectional study by Dibah et al. (26), which has investigated MRSA clinical isolates, noted a high frequency of resistance to clindamycin (94.7%), ciprofloxacin (68.4%), and oxacillin (89.5%). The study by Arabestani et al. (27) also reported high rates of resistance to erythromycin (74.9%) and clindamycin (50.2%) among MRSA isolates, which are consistent with the results of the current study. Nourbakhsh et al. (23) have investigated 110 S. aureus clinical isolates and reported a relatively high prevalence of resistance to clindamycin (54%) (23). Alli et al. (24), in a study conducted in Nigeria, reported resistance rates of 49.4% and 25% for erythromycin and clindamycin, respectively.
An Indian study on S. aureus isolated from bovine raw milk reported resistance rates of 50% and 40% for oxacillin and ciprofloxacin among S. aureus isolates, respectively (28). In another study conducted by Amiri and Anvari (10) on clinical isolates of S. aureus in Rasht Hospital, the rate of resistance to oxacillin was found to be 44.7%. It worth noting that resistance rates reported in the current study are lower than values reported by all the aforementioned studies. This discrepancy could be attributed to the samples, geographical area, study design, and population, dissemination of specific types among patients, and unrestricted policies in consumption and prescription of antibiotics. The influences of policies on infection control, differences in the quality of provided services in hospitals, the emergence of various mechanisms of resistance, and the genetic backgrounds of strains should not be overlooked.
Although the presence of biofilm encoding genes is not always associated with biofilm formability, several studies have mentioned various factors that contribute to biofilm formation and its development in S. aureus isolates, including surface adhesion characteristics, environmental conditions, and genetic backgrounds of the bacteria (4, 9). The association between virulence factors, especially MSCRAMMs, and antibiotic resistance patterns, is well-established in various studies (29-31). Based on the findings, cna, clfB, clfA, fnbA, ebp, and fnbB genes were found in 74.7%, 54.2%, 50.6%, 42.1%, 13.2%, and 12% of isolates, respectively. Similarly, Amini and Mahmoudi-Kojedi (32) investigated the biofilm formation of S. aureus isolated from fresh milk and reported a frequency of 11.6% and 25% for ebp and bbp genes, respectively. Also, cna, clfB, and clfA were the most frequently found biofilm-related genes, which is consistent with studies by Gowrishankar et al. (33) and Duran et al. (34), that reported high prevalence of clfA and cna genes. Ahangari et al. (19) also investigated 75 S. aureus samples isolated from cow mastitis, and reported that 97.3%, 97.3%, 86.7%, 84%, and 84% of isolates were positive for the presence of ebps, fnb, bbp, clfA, and clfB genes, respectively.
Sharma et al. (28) also reported a frequency of 97%, 93%, 90%, and 80% for fnbA, clfB, ebps, and fnbB encoding genes, respectively. Shahmoradi et al. (35) investigated 54 S. aureus strains isolated from urine, blood, mucus, cerebrospinal fluid, pleural fluid, and wounds, and reported the frequency of fnbB (26%) and fnbA (70%) genes. A study that analyzed 123 MRSA isolates from different clinical samples in Hamedan reported that 6.9%, 4.8%, and 13.1% of isolates were positive for bbp, cna, and ebps genes, respectively, which are lower than values reported in the present study (13). Consistent with findings of a study that investigated the role of the bap gene in biofilm production and reported a low prevalence for it (33), in the present study also bap gene was found in 13.2% of isolates. Paniagua-Contreras et al. (36) investigated 109 S. aureus isolates recovered from hemodialysis patients’ catheters in Mexico and could found clfB, clfA, bbp, ebps, cna, and fnbB genes in 81.8%, 100%, 78.2%, 85.4%, 78.1%, and 56.3% of isolates, respectively. Which are higher than the values reported in the current study, except for the fnbA gene, which was found in 34.5% of isolates. These controversies may be due to genetic makeup, gene regulator system, environmental conditions, or type of S. aureus isolates (animal and human).
5.1. Conclusions
The current research presented updated data on the phenotypic resistance and genes involved in the biofilm production of S. aureus isolated from burn patients hospitalized in Tehran, Iran. Based on the findings, resistance to antimicrobial agents and biofilm genes, which often lead to treatment failures and persistent infections, was high. As antibiotic resistance is directly associated with antibiotic use, policies on prescription, and consumption of antibiotics, especially for treating burn patients, need revision. Also, a significant association was found between biofilm encoding genes and reduced susceptibility to antibiotics, which highlights the particular importance of programs for prevention and control of infections as well as precautionary measures to stop the dissemination of these isolates in burn units. However, further studies are needed to investigate the epidemiology of S. aureus, genetic characteristics, and biofilm management upon infection in burn centers.