1. Background
Cryptosporidiosis is considered to be one of the most important diarrheal diseases to humans and many vertebrate animals (1, 2). Exposure to low doses of Cryptosporidium oocysts can cause disease, so it has major importance in public health (3). The parasites’ oocyst is highly resistant to chlorination and disinfectants, which can survive for a long time in the environment (3, 4). It causes up to 6% of diarrhea in immunocompetent individuals (5). Cryptosporidiosis is usually a self-limiting disease; on the other hand, it can be life-threatening in people with immune deficiencies or malnutrition (2).
Medical diagnostic laboratories of Bandar Abbas run routine procedures for the detection of intestinal parasites, but they do not use the proper method for detection of this parasite, unless it is requested by the physician. As performed previously in children with diarrhea in Bandar Abbas, the prevalence of Cryptosporidium spp. was reported as 7% by modified Ziehl-Neelsen (ZN) staining (6). The use of molecular tools in epidemiologic investigations has provided new insights into the diversity of the Cryptosporidium spp. as humans and animal infecting factors (7). The 60-kDa glycoprotein gene (gp60) has a high degree of polymorphism among species isolated from Cryptosporidium and several subgroups, and sub-genotypes have been identified, including Cryptosporidium parvum IIa and IId subtype groups, which are capable of transmission by animals (4). The C. parvum subtype family IIa, preferably infects cattle, whereas IId sheep and goats (8).
2. Objectives
The present study was performed in order to find prevalence and genotypes of Cryptosporidium spp. among patients with diarrhea in Bandar Abbas city, Southern Iran.
3. Methods
3.1. Study Area
In this descriptive cross-sectional study, a single fecal specimen was collected from 170 diarrheic patients in 3 hospitals of Bandar Abbas, Iran, from October 2018 to May 2019. This city is located in southern Iran, a tropical region attached to the Persian Gulf with a high humidity (20% - 100%) and warm climate (9).
3.2. Sample Collection and Processing
After obtaining written consent, the researcher administered a comprehensive questionnaire to each patient in the period of time mentioned above. Recipients of anti-parasitic drugs and diarrheic patients by Shigella spp. were excluded. The checklist included items on patient demographic aspects. Subsequently, a single fecal specimen was collected from 170 diarrheic patients.
3.3. Microscopic Examination
To identify the oocysts of Cryptosporidium spp., a permanent slide was prepared for each sample after the formalin-ether concentration method. Slides stained with the modified ZN- staining were viewed under a light microscope at a final magnification of 1,000 to observe Cryptosporidium oocysts (6).
3.4. DNA Extraction and Nested-PCR
The positive specimens were determined by the staining method, and 47 suspected ones of Cryptosporidium spp, were stored in 2.5% potassium dichromate (K2Cr2O7) and stored at 4ºC for DNA extraction (10). Approximately 200 μL of fecal suspension was washed three times in distilled water before extraction. Genomic DNA was then extracted using the FavorPrep Stool DNA Isolation Mini Kit (FAVORGEN, Taiwan) according to the manufacturer’s instructions. Subtype analysis of Cryptosporidium targeted a gp60 gene fragment (400 bp) using nested PCR as previously described (11, 12) (Table 1).
| Variable | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 |
|---|---|---|---|---|---|---|
| Age | 1 | 3 | 8 | 31 | 32 | 33 |
| Gendera | male | female | female | female | female | female |
| Occupation | - | - | - | housewife | housewife | housewife |
| Educational level | - | - | - | > high school | < high school | > high school |
| Residency | rural | urban | urban | urban | urban | urban |
| Type of reception | inpatient | outpatient | outpatient | inpatient | inpatient | outpatient |
| Contact with animals | yes/sheep | no | no | no | no | no |
| Underlying disease | no | no | no | no | yes/organ transplant | no |
| Travel history in recent 3 months | yes | yes | no | yes | no | yes |
| season | Autumn | Spring | Spring | Winter | Winter | Spring |
| Addict | no | no | no | no | no | no |
| Genotyping | IIdA14G1 | IIdA14G1 | IIdA14G1 | IIdA15 | IIdA14G1 | IIdA14G1 |
aP value = 0.039
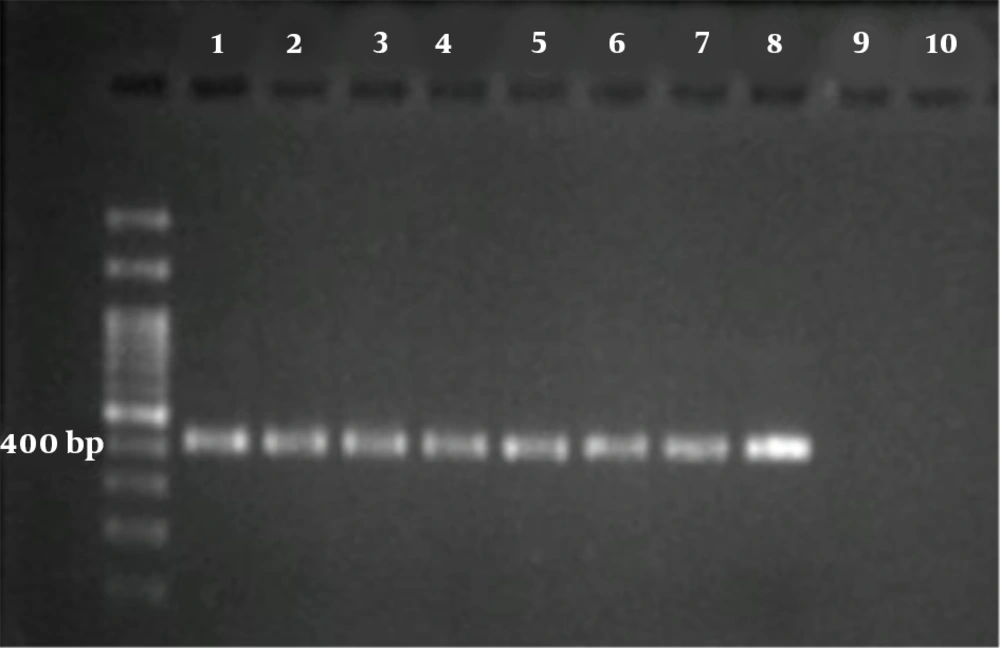
Briefly, in both reactions, the total volume was 20 μL containing 3 μL of MgCl2 solution (25 mM), 2 μL of 10 × reaction buffer, 1.5 μL of 10 mM dNTPs mix, 2 μL of primer mix (10 pm/μL), ~4 ng of DNA template and 0.25 μL of Taq DNA polymerase (5 U/μL) (all from Parstous, Mashhad, Iran). Two PCR cycles were as follows: an initial denaturation at 94ºC for 5 min, followed by 35 cycles of 94ºC for 45 s, 55/58ºC for 1 min/45 s, and 72ºC for 60 s, then final extension at 72ºC for 7 min. For the first reaction, outer primers GP60 forward1 (5’‐ATAGTCTCCGCTGTATTC‐3’) and GP60 reverse1 (5’-GCAGAGGAACCAGCATC-3’) were used at annealing temperature 55ºC, with a product size of 980 - 1,000 bp. For the second reaction, inner primers GP60 forward 2 (5’-TCCGCTGTATTCTCAGCC‐3’) and GP60 reverse 2 (5’-GAGATATATCTTGGTGCG-3’) were used at annealing temperature 58ºC with a product size of nearly 400 bp (Figure 1).
3.5. Sequence Analysis
For final confirmation, PCR products of the gp60 gene (approximately 400 bp) were sequenced on an automated sequencer using primers 5’-TCCGCTGTATTCTCAGCC-3’ and 5’-GAGATATATCTTGGTGCG-3’ (Bioneer Corp). After trimming low-quality sequencing reads at the 5’ and 3’ ends, nucleotide BLAST (Basic Local Alignment Search Tool) similarity searching (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was performed between the PCR-related sequences and sequence databases and the statistical significance was calculated (percent identity). In addition, by using Clustal Omega online software (https://www.ebi.ac.uk/Tools/msa/clustalo), multiple alignment of the trimmed nucleotide sequences (370 bp) was carried out. For enabling classification of C. hominis and C. parvum, as described by Chalmers and colleagues (13), firstly, an allelic family is identified from a conserved sequence of a 3’primer region of the gp60 gene (e.g. IId). Variation in a 5’ trinucleotide repeat region of the gene then identifies subtypes within each family (e.g, A15G1). Finally, in some gp60 families, the number of contiguous copies of a short repeat sequence, Rn, located in a region between two primer regions of the gene, can also contribute to subtype identification (e.g. R1). In this study, in order to recognize isolated subtypes, trinucleotide repeats TCA (red ink), and TCG (green ink) in variable regions located in the 5’primer region of gp60 gene were enumerated (Figure 2).
Multiple sequence alignment of Cryptosporidium parvum isolates obtained from PCR products of gp60 gene. Red and green boxes indicate trinucleotide repeats TCA and TCG in the variable region. Isolates 1, 2, 3, 5, 6 were identical in nucleotides with 14TCA and 1TCG repeats (subtype family IIdA14G1), whereas isolate 4 contained 15TCA repeats without TCG trinucleotide (subtype family IIdA15). The asterisks indicate identical nucleotides.
3.6. Statistical Analysis
The collected data were analyzed using SPSS software (version 20, Chicago, IL, USA), while the relationship between the variables and the presence of Cryptosporidium spp. was assessed by the chi-square test. Frequency (n) and percentage (%) was used to describe qualitative variables. To assess the degree of agreement between PCR and ZN staining for detecting Cryptosporidium spp. Cohen’s kappa-index: poor agreement (k < 0.20), fair agreement (k = 0.21 - 0.40), moderate agreement (k = 0.41 - 0.60), substantial agreement (k = 0.61 - 0.80) and perfect agreement (k = 0.81 - 1.00) was applied (14). For all statistical analyses P < 0.05 was considerate as statistically significant.
4. Results
One hundred and seventy individuals with diarrheic stool were recruited, of which 98 (57.6%) were males and 72 (42.4%) were female. The median age of the study participants was 28.5 yr. (range: 1 d to 91 yr.). Prevalence of Cryptosporidium spp. by modified ZN staining was 1.8% (3/170). However, PCR in positive patients (3 individuals) and 47 suspected specimens revealed infection to Cryptosporidium spp. in 12% (6/50). The youngest infected patient was a one-year-old and the oldest was 33. Evaluating the positive cases of infection, we found that 3 of the cases were housewives.
The results of the chi-square test with the variables showed that the frequency of Cryptosporidium spp. was significantly related only to gender (Table 1). One of the patients had already endured a kidney transplant. Also, 3 of the patients were children and one of them came from the rural area whose parents bred sheep at home. There was no significant difference between age, occupation, education level, residency, type of reception, contact with animals, underlying disease, travel history within the last 3 months, season, and addiction. The other demographic characteristics of the 6 positive patients are presented in Table 1. With regard to Cohen’s kappa-index definition, the agreement level between the two methods, PCR and ZN staining to detect Cryptosporidium spp. was above average (Kappa = 64%). In other words, there is a substantial agreement between the two methods (14). However, PCR detection power was significantly higher than that of ZN staining (P < 0.001) (Table 2).
| PCR | Modified Ziehl Neelsen Stain | Kappa | P Value | ||
|---|---|---|---|---|---|
| Negative | Positive | Total | |||
| Negative | 44 (88) | 0 (0) | 44 (88) | 0.64 | < 0.001 |
| Positive | 3 (6) | 3 (6) | 6 (12) | ||
| Total | 47 (94) | 3 (6) | 50 | ||
4.1. Nucleotide Sequence Accession Number
Nucleotide BLAST similarity results showed that all representative isolates belonged to the C. parvum IId family (13). Isolates 1, 2, 3, 5, 6 had percent identity up to 99.73% with C. parvum IIdA14G1 subtype family under the accession number KT716847.1. Further, isolate 4 exhibited percent identity 97.57% with C. parvum IIdA15G1 subtype family under the accession number HQ241928.1. Multiple sequence alignment of PCR sequences for the gp60 gene illustrated that isolates 1, 2, 3, 5, 6 were identical in nucleotides, whereas isolate 4 revealed a percent identity of 97.84% with the rest. In order to recognize isolate subtypes, trinucleotide repeats TCA (red ink), and TCG (green ink) in variable regions located in the 5’ region of the gp60 gene were enumerated. As a result, Isolates “1, 2, 3, 5, 6” and 4 were assigned as IIdA14G1 and IIdA15 subtype families, respectively (Figure 2). Finally, two distinct nucleotide sequences obtained from this study were deposited in GenBank under the accession numbers MN820454 and MN820453.
5. Discussion
In our study, the infection rate of Cryptosporidium spp. was 1.8%. Prevalence of Cryptosporidium spp. in patients with gastroenteritis in other regions of Iran varies, in Mazandaran province, northern Iran 0.1% (15), Nahavand county in western Iran 1.3% (12), Iranian children of Tehran 2.4% (16), Gonbad Kavoos city, northern Iran 4.94% (17) and in Shiraz, Fars province 25.6% (18). There are differences between our study and a previous one performed in Bandar Abbas (6). This study had less prevalence since our subjects included all individuals with diarrhea, whereas the previous study (6) was performed among children with diarrhea. As a result, the latter had a higher prevalence (7%). Unfortunately, there was no animal study of the prevalence and genotype of Cryptosporidium in Hormozgan to be associated with the results of our study. The prevalence of Cryptosporidium spp. in the other countries of the world also varies. In the rural population of the Buner district, Pakistan, the prevalence of this parasite was found to be 29.88% (19), in Lebanon 11% (20), and in New Zealand 10% (21). This discrepancy may be due to the study population, exposure to animals, residency, geographical climates, nutritional habits, and especially, the type of detection methods (22).
In a systematic review and meta-analysis study in Iran (22), and Lebanon (20), the prevalence of this parasite in children was significantly higher than the other groups, in contrast to our study where there was no significant difference between age and parasitic infection. There found to be a significant difference between the occurrence of infection and gender, consistent with the study of Keshavarz et al. (23) and Khalili and Mardani (24) and inconsistent with the study of Saneian et al. (25). All Cryptosporidium isolates from patients with diarrheal complaints were C. parvum, and none belonged to C. hominis; it indicates transmission of infection from animal to human similar to the study of Sharbatkhori et al. in the northern Iran (17). It is noteworthy that only one patient had direct contact with sheep. Other contamination may have been due to indirect exposure to animal feces, such as polluted vegetables or fruits.
As we can see, all three women are housewives and likely to be infected with dirty vegetables. Most of the infections in Iran are C. parvum (26-28). Molecular studies in the Middle East countries showed C. parvum, as the most dominant species in human infections (22) this is contrary to study of Squire and Ryan, which shows that C. hominis is the most cases of infection in Africa (29) as well as the study of Osman et al. in Lebanon (20) and Sannella et al. in Thailand (30). A few numbers of isolates in the study of Keshavarz et al. (23) in Tehran and Qazvin, Ranjbar et al. and Taghipour et al. in Tehran (16, 28) Rafeie et al. in Ahvaz (10) as well as Mohammadian et al. in Zabol, eastern Iran (27), detected C. hominis while none of the isolates in the present study were C. hominis. In contrary to our study, the other species of Cryptosporidium, C. meleagridis is one of the major human parasitic pathogens in African countries (29).
The gp60 is the most commonly used genetic locus for subtyping Cryptosporidium spp. (8). Nearly 20 C. parvum subtype families have been described at this locus, IIc appears to be adapted to humans, IIa adapted to humans and a broad range of animals, and IId adapted to animals (sheep, goats, and cattle) (31). In this study, sequence analysis of the gp60 locus identified only one C. parvum subtype family, IId, and two subtypes (IIdA14G1 and IIdA15). According to the subtypes found in this study, it appears that the infected individuals are either directly or indirectly in contact with the animal, and the main mode of transmission in Bandar Abbas is zoonotic. One of the animals bred in the rural areas is sheep and goats, which is probably the reason for the high prevalence of this subtype in these areas. Unlike the study of Ranjbar et al. (28) and Sharbatkhori et al. (17) which identified two subtypes (IIa and IId) and Garcia et al. (21) which identified more subtypes (IIa, IIc, IId, and IIe) among the Cryptosporidium isolates, all of the subtypes in the present study were of the IId subtype.
One limitation of the present study was the low number of samples as well as the number of positive samples, but this was the first study to determine the species and genotypes of the parasite in Bandar Abbas. Of course, more molecular studies are suggested to determine the pathways of transmission of this parasite as well as its epidemiology in the wide range of specimens in humans as well as the cattle of Bandar Abbas, Hormozgan province.
5.1. Conclusions
The study confirmed that the transmission of the parasite in Bandar Abbas is more zoonotic than anthroponotic. Therefore, these results are useful for researchers to determine the appropriate preventive and therapeutic measures. In addition, there was a significant difference in parasite detection by microscopic methods compared to molecular methods, so molecular methods are suggested as a more accurate and sensitive methods in cases where we suspect this parasite.