1. Background
Vulvovaginal candidiasis (VVC) is an uncomfortable infection that occures up to 75% of women at least once in their life. Moreover, for a small percentage of women, it occurs more than one episode (1, 2). Due to increased resistance of Candida spp. to current antifungal drugs, the challenge of recurrent vulvovaginal candidiasis (RVVC) is on the rise (2, 3). Both pregnant and non-pregnant women are at risk of vaginal colonization of Candida spp. (4, 5). Candida albicans is the most important cause of VVC, while non-albicans Candida (NAC) species is due to C. glabrata (5, 6). Also, NAC species including C. parapsilosis, C. tropicalis, and C. krusei, are reported as causative agents of VVC, and these species often no longer respond to antimycotic drugs (1, 7).
Furthermore, the difficulty of definitive identification of Candida species using conventional methods has increased, particularly for unusual and closely related species. Amplification of genomic DNA and sequencing of the resulted amplicons is one of the most commonly used molecular diagnostic techniques. Nucleotide sequence assessment of the internal transcribed spacer section of the rRNA gene (8) and the D1/D2 region of the large-subunit rRNA gene are the most widely used techniques for definitive identification of pathogenic fungal species (9, 10).
In the Candida complexes, such as the C. albicans complex, amplification of the hyphal wall protein 1 (HWP1) gene is the appropriate molecular approach for distinguishing C. albicans, C. dubliniensis, and C. africana (11). Various Candida spp. have different levels of susceptibility to antimycotic drugs. Therefore, the exact identification of Candida vaginal isolates is important for providing effective care.
2. Objectives
This study aimed at molecular identification of Candida isolates, causing VVC using nucleotide sequences of the D1/D2 region of the large-subunit rRNA gene.
3. Methods
3.1. Patients and Collecting Isolates
This cross-sectional study was performed on 320 non-pregnant vaginitis patients referred to Shahid Akbar-Abadi Obstetrics and Gynecology Teaching Hospital in Tehran (Iran) from 2016 - 2017. Besides, information on age, occupation, medical history, symptoms, and recurrent episodes (recurrent VVC was defined as ≥ 4 episodes within a year) were also collected (12). Vaginal secretions were collected from the posterior fornix with a sterile cotton swab and put into 3 mL of sterile saline.
3.2. Exclusion Criteria
Infection with Mycoplasma spp., Trichomonas vaginalis, Chlamydia, or any bacterial vaginosis or vulvar skin infection. History of using local or systemic antifungal compounds during the past two weeks.
3.3. Yeast Identification
Microscopic assessment was used to identify yeast forms or pseudomycelium. All specimens were cultured on CHROMagar Candida (CHROMagar Candida, France) for colony count and distinguishing concomitant of Candida spp. infection. The identification was confirmed by germ tube formation, chlamydospore production in cornmeal agar (CMA) (Merck Germany) with 1% Tween 80, and cultured on Sabouraud dextrose agar medium with chloramphenicol at 42°C.
3.4. Molecular Methods
For DNA extraction a single colony on CHROMagar Candida was selected and subcultured on yeast extract peptone dextrose (Merck Germany) (YEPD) agar at 37°C for 24 - 48 h. Genomic DNA of yeast isolates was extracted using the Qiagen DNA tissue kit (Germany) and deposited at -20°C for until use. The universal primers NL1 (5′-GCA TAT CAA TAA GCG GAG GAA AAG-3′) and NL4 (5′-GGT CCG TGT TTC AAG ACG G-3′) were applied for amplification of the D1/D2 region of large subunit ribosomal RNA (10) by the subsequent program: 98°C (5 min), 35 cycles of 98°C (30 s), annealing temperature 60°C (30 s), and 72°C (30 s) and 72°C for a 5-minute final extension. The amplicons were sequenced by Macrogen (Korea). Sequences were evaluated by the reference nucleotide sequences retrieved from the GenBank database with the BLAST sequence search tool (http://www.ncbi.nlm.nih.gov/BLAST), and results were deposited to GenBank. The PCR primers CR-f (5′-GCTACCACTTCAGAATCATCATC-3′) and CR-r (5′-GCACCTTCAGTCGTAGAGACG-3′) amplified the HWP1 gene for identification C. albicans, C. africana, and C. dubliniensis according to Romeo and Criseo PCR condition introducing method (11). Candida albicans (ATCC 10231) was used as the reference strain.
3.5. Phylogenetic Analysis
The sequencing results of the D1/D2 region was examined and compared with the reference nucleotide sequences of Candida spp. using the neighbor-joining method by MEGA 7.
4. Results
In total, 320 non-pregnant vaginitis patients were included in the present study. That 100 patients aged 18 - 56 years showed vaginal candidiasis. Most of them were found in isolates of those aged 30 to 39 years old. 86% and 14% of cases were VVC and RVVC (Table 1). Most of the participants were housewives (27%), followed by office workers (25%), factory workers (22%), and self-employed (26%). None of the participants had underlying diseases like diabetes, urinary tract infections, immunocompromised diseases, or any other chronic diseases. Primary identification of the Candida spp. was conducted by germ tube test, CHROMagar Candida, chlamydospore production, and cultured at 42 ºC. C. albicans produced green-colored colonies on CHROMagar Candida, chlamydospores on CMA medium, and appeared germ tube in the test, as well.
| Patient Characteristics | Candida Species | ||||
|---|---|---|---|---|---|
| C. albicans | C. glabrata | C. krusei | C. kefyr | Totala | |
| Age, y | |||||
| > 20 | 3 | 1 | 0 | 0 | 4 (4) |
| 20 - 29 | 17 | 13 | 3 | 3 | 36 (36) |
| 30 - 39 | 22 | 14 | 4 | 0 | 40 (40) |
| 40 - 49 | 8 | 6 | 1 | 2 | 17 (17) |
| 50 - 59 | 1 | 2 | 0 | 0 | 3 (3) |
| Total | 51 | 36 | 8 | 5 | 100 |
| Vaginal candidiasis | |||||
| VVC | 44 | 32 | 6 | 4 | 86 (86) |
| RVVC | 7 | 4 | 2 | 1 | 14 (14) |
| Total | 51 | 36 | 8 | 5 | 100 |
Abbreviations: 1VVC, vulvovaginal candidiasis; 2RVVC, recurrent vulvovaginal candidiasis.
aValues are expressed as No. (%).
Candida africana generated small green colonies on CHROMagar Candida and did not generate chlamydospores on CMA medium. Candida albicans strains grew at 42°C, while the C. africana isolates did not. Candida glabrata and C. kefyr (Kluyveromyces marxianus) produced dark pink colonies with pale edges and light violet colonies respectively. The clinical isolates with green-colored colonies on CHROMagar Candida and reference strain were analyzed with CR-f and CR-r specific primers used to amplify the HWP1 gene, and the amplification of the ~900-bp fragment confirmed C. albicans isolates (Figure 1).
The PCR of the D1/D2 region of the large-subunit rRNA gene with NL1 and NL4 primers amplified ~500 bp fragment (Figure 2). The D1/D2 nucleotide sequences of Candida spp. clinical strains were analyzed with reference nucleotide sequences retrieved from the GenBank database using BLAST. The species of all clinical isolates were identified. The D1/D2 sequences of Candida spp. clinical isolates were deposited in GenBank under accession numbers KY548531-2, KY548534-KY548548, KY548550, MG025904-MG025928, MG025930-MG025933, MG966211-MG966257, and MG967349-MG967353. C. albicans to be the most strain (51%) in total of VVC patients, followed by C. glabrata (36%), C. krusei (Pichia kudriavzevii) (8%), and C. kefyr (K. marxianus) (5%). Among 14 RVVC cases, the most common pathogen was C. albicans (50%), followed by C. glabrata (28.6%), C. krusei (P. kudriavzevii) (14.3%), and C. kefyr (K. marxianus) (7.1%) (Table 1). The nucleotide sequences of the D1/D2 region were aligned for phylogenetic evaluation. Candida albicans isolates showed similarity with KU729162 (C. albicans strain ATCC 28121), KY825125.1 (C. albicans isolate DA46), and MG859668.1 (C. albicans isolate HCM-NM58) reference strains. Candida glabrata strains matched completely with MG859667.1 (C. glabrata isolate HCM-NM56) and MG228366.1 (C. glabrata isolate DMic 154894) reference strains in gene bank databases. Candida krusei (P. kudriavzevii) indicated 100% similarity to MG859665.1 (HN-NM19 isolate), C. kefyr (K. marxianus) displayed 100% identity with MH595097.1 (UCDFST: 49-27 isolate) and CP023460 reference strain (Figure 3).
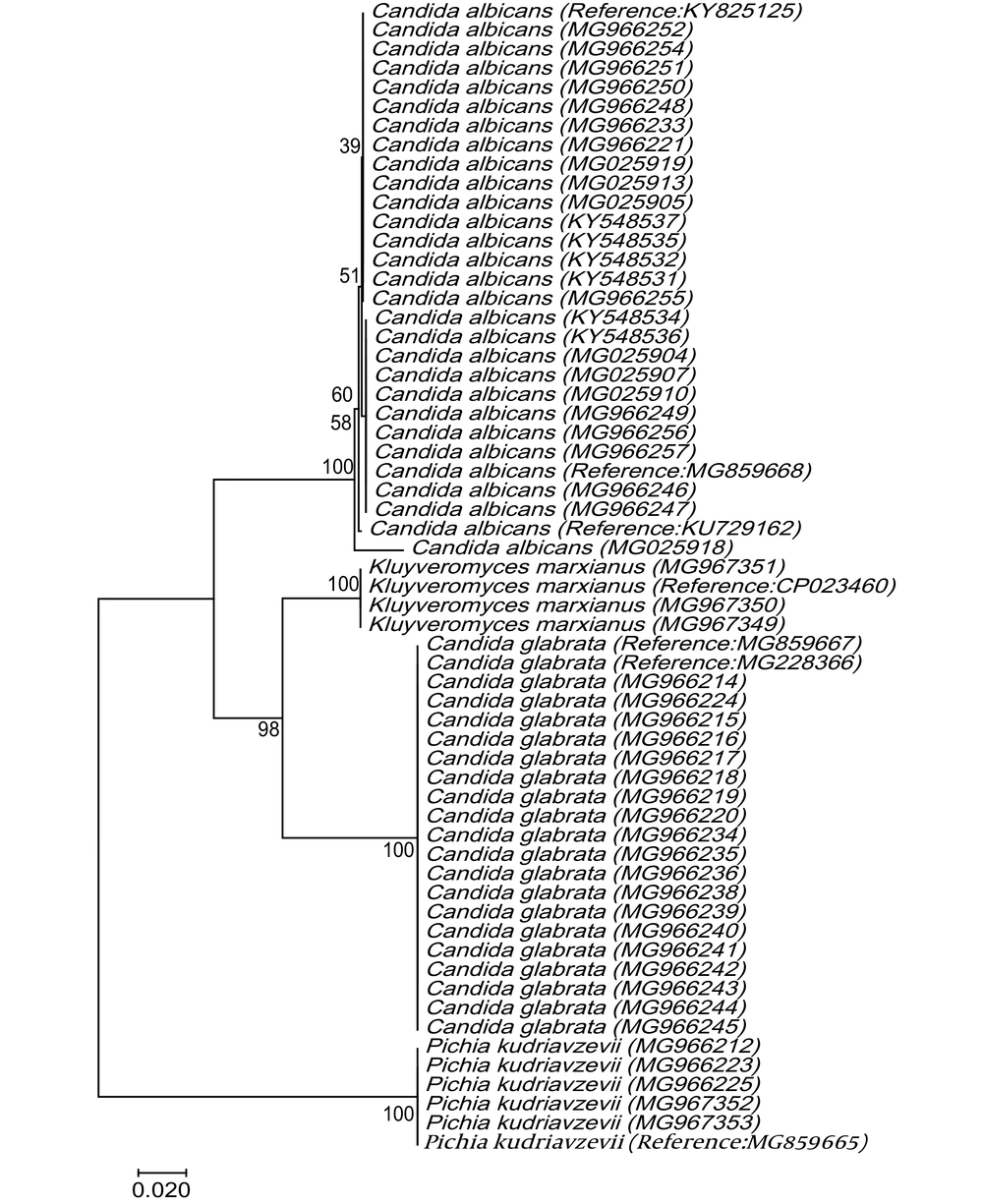
The phylogenetic tree of Candida spp. was inferred based on the nucleotide sequences of the D1/D2 region. The evolutionary relationship of Candida spp created by the neighbor-joining method, based on the nucleotide sequences of D1/D2 region retrieved from this study (KY548531-KY548532, KY548534-KY548537, MG025904-MG025905, MG025907, MG025910, MG025913, MG025918-19, MG966212, MG966214-MG966221, MG966223- MG966225, MG966233-MG966252, MG966254-MG966257, MG967349-MG967353) compared with reference sequences of C. albicans strains (KU729162, KY825125 and MG859668), C. glabrata strains (MG859667 and MG228366), C. krusei (Pichia. kudriavzevii; MG859665), and C. kefyr (Kluyveromyces marxianus; CP023460) from GenBank. Bootstrap values achieved from 1000 replicates are indicated on branches in percentage. Evolutionary evaluates were presented in MEGA7.
5. Discussion
In the present study, Candida spp. was the most important agent of the fungal vaginitis. That is in line with the findings of the previous studies that reported C. albicans and C. glabrata as the most frequent pathogenic yeasts in VVC patients (13-15). Several studies reported that the percentage of NAC isolates in vaginitis (5, 16) and also infections caused by unusual yeasts is on the rise (17). Traditional methods cause difficulties for definitive diagnosis of the multiplicity of yeast species. Molecular approaches can improve the differentiation of less common yeast isolates and strongly similar yeast species such as those in Candida complexes. DNA based approaches are most valuable for large epidemiological studies on unusual yeasts. Although it rarely occurs, several studies have reported the Saccharomyces cerevisiae as the agent of genitourinary infection. In the present study, we couldn’t find this yeast. However, since it’s less susceptible to the currently available antifungal drugs, S. cerevisiae is an important pathogenic yeast (18, 19).
In this study, routine mycological methods and DNA sequencing were used to identify yeast isolates. It’s proved that sequencing the D1/D2 or ITS regions is an accurate approach for definitive identification of pathogenic yeast species, particularly the C. glabrata (17, 20), and the C. parapsilosis complexes (21). The phylogenetic tree was built using the sequences of the D1/D2 region of different Candida strains and indicated the creation of distinct branches for Candida species. The phylogenetic analyses of the D1/D2 region showed that all C. albicans isolates had 100% homology with KU729162 (C. albicans strain ATCC 28121), KY825125.1 (C. albicans isolate DA46), and MG859668.1 (C. albicans isolate HCM-NM58) reference strains. C. glabrata strains matched completely with MG859667.1 and MG228366.1 reference strains in gene bank databases. C. krusei (P. kudriavzevii) showed 100% homology with MG859665.1, and C. kefyr (K. marxianus) showed 100% identity with MH595097.1 and CP023460 reference strains.
Hasanvand et al. (7) investigated C. albicans, C. glabrata, C. tropicalis, and C. parapsilosis isolated from vaginitis patients and reported C. albicans as the main cause of vaginal candidiasis. Several studies conducted in Iran and other countries reported C. albicans as the predominant pathogen in VVC (2-4, 6, 7, 12-16, 18, 22-24). Another study has reported C. tropicalis as the main NAC species that cause VVC (25), which is not consistent with the current study that found C. glabrata as the first most frequently encountered NAC species.
Several studies mentioned C. glabrata as the important NAC species in Candida vaginitis patients (5, 12, 14). The current study revealed that C. krusei (P. kudriavzevii) and C. kefyr (K. marxianus), as NAC species, are associated with vulvovaginitis. Clinical isolates like C. krusei and C. glabrata were resistant or showed low susceptibility to azole drugs, as reported by other studies (5, 18). Candida glabrata is reported as a common fungal pathogen in many countries and appears to be intrinsically resistant to fluconazole.
Another study reported C. lusitaniae isolates as the second most common Candida species have also shown susceptible to all tested antifungal drugs in VVC patients (24), although this yeast was not reported in the current study. Other NAC species such as C. tropicalis, C. parapsilosis, Meyerozyma guilliermondii, C. dubliniensis, C. famata, C. africana, and C. orthopsilosis have frequently been reported from VVC patients (7, 22, 24). In this study, C. albicans and C. glabrata were the most important agents in VVC and RVVC.
Candida kefyr (K. marxianus) and C. krusei (P. kudriavzevii) were isolated from VVC and RVVC patients, as well. In the present study VVC and RVVC patients were healthy women without a history of exposure to known risk factors such as diabetes, pregnancy, immunodeficiency disorders, or any chronic disease. According to the clinical studies, most of RVVC patients are healthy women, and multiple recurring infections are often idiopathic without known potential risk factors (26). Numerous studies reported C. albicans as the most prevalent species, even more than NAC species, in vaginal candidiasis patients (2, 6, 7, 13, 15, 18, 22). Overall, C. albicans and non-albicans species had almost similar rates of prevalence (51% versus 49% respectively). Since NAC species often can be find in RVVC patients and do not respond to standard antifungal therapies, their identification is of crucial importance. The results of this study indicated that C. albicans and C. glabrata are still the principal pathogenic agents in recurrent and non-recurrent Candidal vaginitis.
5.1. Conclusions
Candida albicans and C. glabrata are the most important Candida species isolated from VVC and RVVC, which confirms the dramatic incidence of NAC species in VVC patients. Accurate identification of Candida spp. is important for treatment administration because of some strains revealed different levels of resistance to antimycotic medicines.