1. Background
Bloodstream infection (BSI) is one of the life-threatening diseases with one-year mortality ranging from 8 to 48% (1). It manifests as changes in temperature (> 38°C or < 36°C), tachycardia (> 90 per minute), or peripheral white blood cell count (< 4000 or > 12,000 cells/µL or > 10% of immature forms of granulocytes [stripes]) (2). However, timely and correct diagnosis can significantly improve outcomes. Every hour of delay in antimicrobial administration within the next six hours is associated with an average 7.6% decline in survival (3). Therefore, the correct detection of pathogenic bacteria is essential for guiding the treatment with antibiotics. Rhodes et al. (4) recommended that appropriate routine microbial (including blood) cultures should be obtained before starting antimicrobial therapy in patients suspected of sepsis or septic shock.
Blood cultures, however, are time-consuming and slow. On the other hand, they only detect surviving microorganisms and show lower sensitivity to slow-growing intracellular and difficult microorganisms. In addition, despite the correct implementation of standard procedures, the collection of sufficient blood, and a high degree of suspicion of BSI, the overall positive rate of blood culture tests may be as low as 30 - 40% (5). Therefore, it is critical to find a new test method to guide us in the accurate use of antibiotics. The goal of our work was to develop an alternative diagnostic platform for identifying infectious microorganisms based on unbiased sequence analysis of circulating cell-free DNA (cfDNA) in plasma samples of patients with sepsis based on next-generation sequencing (NGS) (6). It is a technology that can replace many traditional microbial workflows and provide clinicians and public health experts with more applicable information than has been achieved so far (7).
2. Objectives
In this paper, 402 plasma specimens from suspected BSI patients in a hospital were collected, and the blood cultures and NGS were analyzed. We hope to use this new technology to assist in the diagnosis and treatment of BSI patients.
3. Methods
3.1. Sample Collection
The data of 63 patients were collected who were considered to have BSI by clinicians in the First Affiliated Hospital of Kunming Medical University from July 31, 2018, to February 28, 2019.
3.2. Next-Generation Sequencing Method
The next-generation sequencing method was done by the Illumina NextSeq 550 Sequencing platform. We verified the existence of microorganisms in patients according to the following principles: a) Positive bacteria, fungi, and viruses: Sequencing number ≥ 10; b) Weakly positive: 3 ≤ sequencing number < 10; and c) Negative: sequencing number < 3 or no detectable pathogen.
3.3. Blood Culture
Blood samples were collected by an attending clinician into blood culture bottles for adults, and the blood culture was performed at a clinical microbiology laboratory. In the present study, we defined the reporting time of blood culture results as the time between the collection of blood samples and reporting to the attending physician by the laboratory, which was consistent with daily clinical activities. A total of 339 bottles of blood culture were evaluated.
3.4. Statistical Analysis
The positive predictive value (PPV), negative predictive value (NPV), sensitivity (SEN), specificity (SPE), and Youden index were obtained by drawing a quadruple table. The diagnostic consistency test and McNemar’s test (a quadruple table for matching) were used to compare NGS and blood culture results with a significance level of 5%. Statistical analyses were conducted using SPSS 22.0.
4. Results
4.1. Description of the Study Participants
A total of 339 specimens for blood culture and 63 samples for NGS were included in the study. The majority of the samples were obtained from patients aged 48 - 67 (52.4%), and 71.4% of them were males. Patients from the Intensive Care Unit, Emergency Care Unit, Surgery Ward, and Internal Medicine Ward accounted for 18 (28.6%), six (9.5%), 27 (42.9%), and 12 (19%) patients, respectively. The infection sites of these patients were considered to be the central nervous system, lung, abdomen, urinary tract, and skin. Among 63 patients, the fever rate was as high as 95%, and 42 (66.7%) patients had a fever above 39°C. A total of six (9.5%) patients died during the study period (Table 1).
| Sample | |
|---|---|
| Total | 63 |
| Age, median (IQR) | 56 (48 - 67) |
| Gender, male | 45 (71.4) |
| Intensive Care Unit | 18 (28.6) |
| Surgical Department | 6 (9.5) |
| Medical Department | 27 (42.9) |
| Emergency Care Unit | 12 (19) |
| Septic focus | |
| Central nervous system | 3 (4.8) |
| Lung | 33 (52.4) |
| Abdomen | 18 (28.5) |
| Urinary tract | 6 (9.5) |
| Skin | 3 (4.8) |
| Fever | 60 (95) |
| < 37.3 | 3 (4.7) |
| 37.3 - 38 | 3 (4.7) |
| 38 - 39 | 15 (23.8) |
| > 39 | 42 (66.7) |
| First blood routine examination on admission | |
| WBC | 63 |
| 4 - 10 × 109/L | 33 (52.4) |
| > 10 × 109/L | 30 (47.6) |
| N, % | 63 |
| < 50 | 6 (9.5) |
| 50 - 75 | 9 (14.3) |
| > 75 | 48 (76.2) |
| Lymph | 63 |
| < 0.8 × 109/L | 0 |
| 0.8 ~ 4 × 109/L | 12 (19) |
| > 4 × 109/L | 51 (81) |
| Index of infection | |
| PCT (procalcitonin), ng/mL | 39 |
| < 0.5 | 15 (38.5) |
| 0.5 - 2 | 15 (38.5) |
| 2 - 10 | 3 (7.7) |
| > 10 | 6 (15.3) |
| CRP (C-reaction protein), mg/L | 39 |
| < 10 | 12 (30.8) |
| > 10 | 27 (69.2) |
| ESR (erythrocyte sedimentation rate), mm/h | 30 |
| < 20 | 6 (20) |
| > 20 | 24 (80) |
| Antimicrobial therapy | |
| Oxazolidone | 8 |
| Tetracycline | 5 |
| Quinolone | 8 |
| Glycopeptide | 6 |
| β-lactam | 37 |
| Antifungal therapy | 10 |
| Antiviral therapy | 8 |
| Number of sample tests | |
| NGS | 63 |
| Blood culture (mean) (CI) | 339 (5) (4.3 - 6.5) |
| Duration of inspection, h | |
| NGS (range) | 18h (16.6 - 20.4) |
| Blood culture (range) | 96h (64 - 120) |
| Outcome | |
| Improvement | 39 (61.9) |
| Requesting discharge | 18 (25.6) |
| Death | 6 (9.5) |
aValues are expressed as No. (%) or median (range).
4.2. Analysis of Clinical Data
According to fourfold Table 2, we calculated SEN, SPE, PPV, NPV, and Youden index as 100%, 5.9%, 20%, 100%, and 5.8%, respectively. Significant differences were observed in the detection of infections between blood culture and NGS methods (McNemar’s = 46; P< 0.01). The positive rates of blood culture and NGS were 23.8% and 95%, respectively [95% CI: 13.3% - 34.3% vs. 95% CI: 90% - 100%].
| NGS | Blood Culture | Total | |
|---|---|---|---|
| + | - | ||
| + | 12 | 48 | 60 |
| - | 0 | 3 | 3 |
| Total | 12 | 51 | 63 |
4.3. NGS Had a Larger Detection Range Than Blood Culture
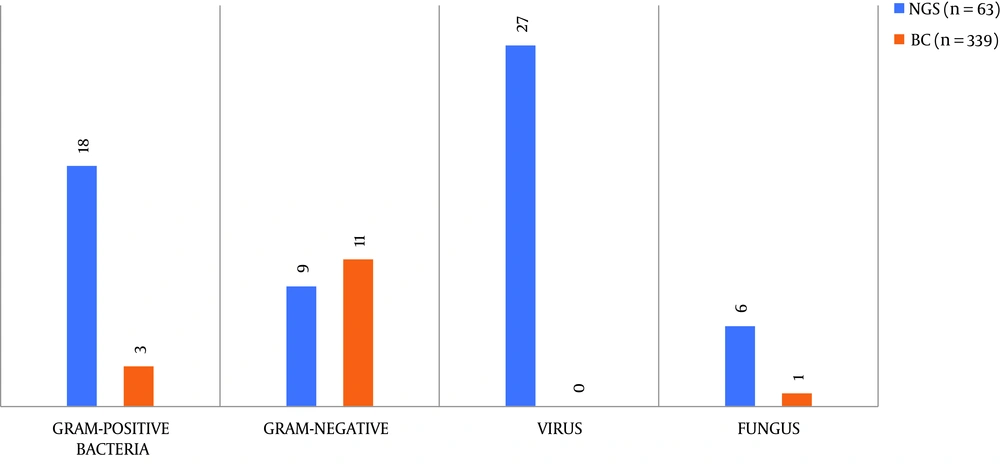
A total of 19 pathogen types, including bacteria, fungi, and viruses, were detected from 63 samples of NGS and 339 samples of blood culture. From Figure 1, we can see that the pathogens detected by NGS in 63 blood samples included Gram-positive bacteria (28.6%), Gram-negative bacteria (14.3%), fungi (9.6%), and viruses (42.3%), while blood culture only detected Gram-positive bacteria (0.8%), Gram-negative bacteria (3.2%), and fungi (0.3%) in 339 blood samples (P < 0.05). It is worth noting that five microorganisms were detected in two specimens/person by blood culture. Therefore, we can conclude that the detection range of NGS was significantly larger than that of blood culture. Supplementary Table 3 lists the detailed results of samples detected by the two different methods.
| Microorganism | NGS | Blood Culture |
|---|---|---|
| Gram-positive bacteria | 18 | 3 |
| Staphylococcus epidermidis | 7 | 0 |
| S. genera | 8 | 0 |
| Micrococcus luteus | 2 | 0 |
| Streptococcus sanguis | 0 | 3 |
| Bifidobacterium breve | 1 | 0 |
| Gram-negative bacteria | 9 | 11 |
| Fusobacterium nucleus | 4 | 0 |
| Pseudomonas aeruginosa | 4 | 0 |
| Neisseria meningitidis | 1 | 0 |
| Stenotrophomonas maltophilia | 0 | 1 |
| Pseudomonas aeruginosa | 0 | 3 |
| Klebsiella pneumoniaepneumonia subspecies | 0 | 7 |
| Viruses | 27 | 0 |
| Human herpesvirus 1 | 2 | 0 |
| Parvovirus | 17 | 0 |
| WU polyomavirus | 4 | 0 |
| Epstein-Barr virus | 3 | 0 |
| Human cytomegalovirus | 1 | 0 |
| Fungi | 6 | 1 |
| Candida parapsilosis | 1 | 0 |
| Yarrowia lipolytica | 5 | 0 |
| Cryptococcus Neoformans | 0 | 1 |
| Unrecognizable pathogenic microorganisms | 3 | 324 |
| Total | 63 | 339 |
4.4. Difference in the Number of Blood Culture and NGS Tests
We collected 339 blood cultures from 63 patients during hospitalization for blood culture detection, and the pathogenic microorganisms were detected in 15 samples, showing a positive rate of 4.4%. However, NGS detected the pathogens in 60 of 63 blood samples, suggesting a positive rate of 95.2%. There was a statistical difference in the positive rate between blood culture and NGS (P = 0.03). A total of 339 blood cultures were performed in 63 patients, with an average of five blood cultures per patient (CI 4.3 - 6.5).
4.5. Next-generation Sequencing Had Much Shorter Detection Time Than Blood Culture
It usually takes 96 h (95% CI: 64 - 120) for a frontline clinician to obtain the results from collecting a patient’s blood sample using the blood culture method. However, the NGS method could obtain and send the results to the doctors’ computer after only 18 h (95% CI: 16.6 - 20.4) of collecting the blood sample.
5. Discussion
Bloodstream infections not only are life-threatening but also increase mortality, morbidity, and health care costs (8). The purpose of our study was to compare the clinical advantages and disadvantages of blood culture and NGS. The results of the current study illustrated that the NGS technology is superior to the blood culture method for the simultaneous diagnosis of different types of pathogens. When all pathogenic microorganisms (e.g., bacteria, fungi, and viruses) were considered in the sufferer, the detectability improved significantly from 4.4% (15/339) to 95.2% (60/63). This marked difference in the detection of all pathogens between the two methods increases the possibility that NSG has higher susceptible to patients with BSI than blood culture than blood culture. However, the NGS diagnosis of clinical pathogens has a long way to go before it can truly benefit patients.
Yet, the existence of diverse bacterial communities in the blood of patients with BSI remains an open question. Blood culture is considered to be the gold standard for the diagnosis of bacteria, but it has the inherent disadvantage of being unable to diagnose viruses and bacteria that are difficult to cultivate. In contrast, NGS technology shows its excellence in virus diagnosis and does not require knowledge of the pathogen’s sequence before detection (9). Within six hours after the consultation, every hour of delay in antimicrobial administration reduces survival by an average of more than 7% (2). However, with NGS technology, we will be able to obtain more information concerning patients with severe infections and do not need to wait 3 - 8 days. Therefore, although it is not recommended that doctors perform appropriate tests as soon as possible, it is encouraged to collect more data from the general population of sepsis patients to confirm the diagnosis without a specific framework. Patients suspected of sepsis and shock should be treated immediately with antibiotics, as there is no room for error (10).
During sample collection, we noticed that a severely infected patient usually needed to do more than five blood cultures during the hospital stay, and a single blood culture usually costs 200 Yuan, so blood cultures during the hospital stay consume a lot of manpower, time, and money. At the same time, proper routine microbial culture should include at least two blood cultures (aerobic and anaerobic) (4). With the technical and monetary barriers to NGS lowered, NGS should be applied to some severely infected patients. Although there are well-documented challenges associated with blood culture, ranging from pre-analytical (antibiotic treatment before collection, ensuring correct collection volume, avoiding contamination) to analytical ones, continuous monitoring of blood culture systems will continue to be the mainstay of BSI diagnosis (11). There are only sporadic reports on the application of NGS in the analysis of clinical samples, although the emerging NGS technique becomes increasingly important in clinical microbiology. Thus, in the future, we need more samples to evaluate the pathogenic, diagnostic effect of NGS on patients with early severe infections.
Initially, NGS was mainly used for research purposes, but as costs decreased, automation, detection sensitivity, library preparation, and sequencing technology options increased, and published guidelines provided standardization, so the practicality of NGS in clinical laboratories was proven (12). We believe that this trend may continue, thereby promoting the wide application of NGS in clinic practice, not just in the diagnosis of pathogens. At a time when many people believe that the post-genomics era has arrived, next-generation sequencing has proven to have great potential for anyone in the field of life sciences (13). Besides, in this COVID-19 pandemic, NGS has contributed to the detection of pathogens. At the same time, we are aware that the research is slightly innovative but may also contribute to the promotion of clinical NGS.
Despite the limited number of samples, this study confirms the great potential of NGS combined with traditional blood culture in the diagnose of BSI and shows its effect on improving the microbiology of pathogen-directed therapy.
5.1. Conclusions
Our results suggest that the NGS method may provide a new diagnostic tool for patients with BSI. Its broad testing range, high positive rate, and rapid detection will benefit BSI patients.