1. Background
Streptococcus pyogenes or Group A streptococcus (GAS) is otherwise known as a “flesh-eating bug” that is one of the major human pathogens responsible for over 600 million infections and about 517, 000 deaths annually (1, 2). The ability of this pathogen to produce an array of deadly virulence factors resulted in heightened concerns among scientists and physicians. One such virulence factor is known as streptococcal pyrogenic exotoxin B (speB), a powerful cysteine proteinase, which is predominantly found in GAS and is involved in GAS pathogenesis (3). Unlike nosocomial threats caused by multidrug-resistant pathogens, notably methicillin-resistant S. aureus, vancomycin-resistant enterococci, and other nosocomial Gram-negative bacteria, GAS diseases are frequently neglected in Malaysia (4, 5). To date, there were no population-based studies conducted on the incidence of GAS diseases such as acute renal failure (ARF) or chronic rheumatic heart disease (CRHD) in Malaysia (6). Ironically, the number of CRHD patients who require mitral valve repair has increased to almost 190 cases per year (7).
The speB gene is predominantly found in almost all GAS isolates. Thus it is considered an important biomarker for GAS identification (8, 9). Since S. pyogenes is still susceptible to penicillin, it is important to diagnose the onset of infection early to reduce the inflammation and prevent the spread of this bacterium (10). The culture method is considered a gold standard, and conventional PCR is commonly used for bacterial detection. However, these traditional methods are often time-demanding and expensive (11-13).
2. Objectives
The loop-mediated isothermal amplification (LAMP) method has recently been accepted as a novel method for the detection of pathogens, including GAS for its rapidity, high sensitivity, and specificity (14). However, a simple, rapid, and highly sensitive technique for measuring the time to positivity of speB gene amplification products in GAS by LAMP real-time turbidimetry method is still lacking. Thus, this study aimed to improve the visual detection of the speB gene by real-monitoring the amplification product by a turbidimetry method in RT-LAMP assay that can be a frontier in the point of care testing for the detection of S. pyogenes in hospitals.
3. Methods
3.1. Sample Collection and Bacterial Identification
A total of 43 bacterial isolates consisting of S. pyogenes clinical strains were used to develop the RT-LAMP assays. These clinical strains were previously isolated from clinical specimens such as pus, blood, wound, tissue, and throat swab (15). Moreover, S. pyogenes ATCC 19615 was used as a positive control in this study. Isolates stored at -70°C in a Luria-Bertani broth (Difco Laboratories, USA) supplemented with 20% glycerol isolates were thawed accordingly for further use. The identification of each clinical strain was conducted by using phenotypic and genotypic methods such as biochemical tests and 16s rRNA.
3.2. DNA Extraction
A single colony was picked from the solid culture medium grown from 5% sheep blood Muller Hinton agar (Isolab Bhd, Sdn, Malaysia) following 24-h incubation under 5% CO2. The colony was transferred into an eppendorf (EP) tube containing 50 µL of double-distilled water. The EP tube was then boiled for 8 - 10 min at 100°C in a heating block (Yinzhou Company, China) and was allowed to cool on ice for 2 min. Following the cooling, the EP tube was centrifuged at 13,000 × g for 5 min. The supernatant containing the genomic DNA was stored at 4°C and used as a template for the RT-LAMP development.
3.3. PCR Assay for Preliminary Detection of Streptococcal Pyrogenic Exotoxin B (speB) Gene
Primers established by Cao et al. were used for the preliminary screening of speB gene detection (16). The PCR assays of 43 S. pyogenes clinical isolates were performed by using DNA Mini Kit (QIAGEN Company, Germany). The total volume of 25 µL reaction mixture contained the following components: 12.5 µL of 2 x EconoTaq® PLUS GREEN master mix [Tiangen Biotech (Beijing) Co., Ltd], 0.5 µL forward (F3) and reverse (B3) primers, 1 µL of sample DNA, and 10.5 µL of distilled water. The PCR cycling conditions were as follows: 94°C for 4 min, followed by 30 PCR cycles of 94°C for 1 min (DNA initial denaturation), 58°C for 1 min (annealing of primer), and 72°C for 1 min (DNA extension), and a final extension at 72°C for 10 min. All PCR products were analyzed by 0.84% agarose gel electrophoresis and visualized using an ultraviolet transillumination light cabinet (Alpha Imager TM 2200, Alpha Innotech, Australia).
3.4. Optimization of LAMP Assay for the Detection of speB Gene
To rule out false-positive amplification, a total of 25 μL reaction mixture containing 12.5 μL of 2 X Reaction Mix [Tris-HCl 40mM (pH 8.8), KCl 20 mM, MgSO4 16 mM, (HN4)2SO4 20 mM, Tween20 0.2 wt%, betaine 1.6 M, dNTPs 2.8 mM], 1 μL (8 U) of Bacillus stearothermophilus (Bst) DNA polymerase, and 1 μL of fluorescent dye (calcein) (Eiken Co., Ltd., Japan) was added into the mixture. In addition, each of FIP and BIP, 1 μL of LB, and finally 1 μL of standardized DNA and 4.5 μL deionized water was also added. Each RT-LAMP reaction was carried out in a turbidimeter (LA-500, Eiken Company, Japan) at different temperatures ranging from 60°C to 65°C at 1°C interval during the optimization of the RT-LAMP assay. The turbidity was measured at 650 nm. The LDR photo-detector captured the light that passed through the reaction tubes, and the signals were measured by real-time signals as sigmoidal graphs. The optimization process also involved different amplification times (10 to 60 min). The reaction product was also visualized by observing color changes (from orange to green) through the naked eye, as well as green fluorescence under UV light (17). Finally, the PCR products were analyzed using 1.5% agarose gel electrophoresis.
3.5. Specificity and Sensitivity of speB Detection by the RT-LAMP Assay
To evaluate the specificity of the RT-LAMP assay for speB gene detection, the genomic DNA of the 11 strains was amplified using the optimized RT-LAMP protocol described earlier. The sensitivity of the RT-LAMP assay for speB gene detection was evaluated using 10-fold serial dilutions of the total genomic DNA of S. pyogenes ATCC 19615. The genomic DNA concentration was measured using a Micro-Spectrophotometer (Nanodrop ND- 1000, Bio-Rad, USA) and was adjusted to 100 ng/μL with sterile distilled water. This was followed by serial dilution from 102 ng/μL to 10-6 ng/ μL based on the established protocols (16). In addition, similar steps were performed for the PCR assay. A total of 11 bacterial strains were used for the specificity in the present study as follows: Streptococcus pyogenes ATCC 19615, Streptococcus pyogenes (clinical isolate), Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, Pseudomonas aeruginosa ATCC 27853, Streptococcus pneumoniae ATCC 6305, Staphylococcus aureus ATCC 33592, Streptococcus agalactiae NCTC 8017, Staphylococcus epidermidis ATCC 12228, Escherichia coli (clinical isolate), and Enterococcus faecalis (clinical isolate).
3.6. RT-LAMP Validation and DNA Sequencing
The RT-LAMP assay for speB gene detection was validated using the genomic DNA extracted from 43 S. pyogenes clinical isolates. While S. pyogenes ATCC 19615 and DDW were used as positive and negative controls, respectively. The LAMP products were purified using QIAEX1 II Gel Extraction Kit (QIAGEN, Shanghai, China) according to the established protocol described by Walker et al. and were dispatched to First Base Sdn. Bhd. (Malaysia) for sequencing (18).
4. Results
4.1. Evaluation of RT-LAMP Assay at Different Temperatures and Reaction Times
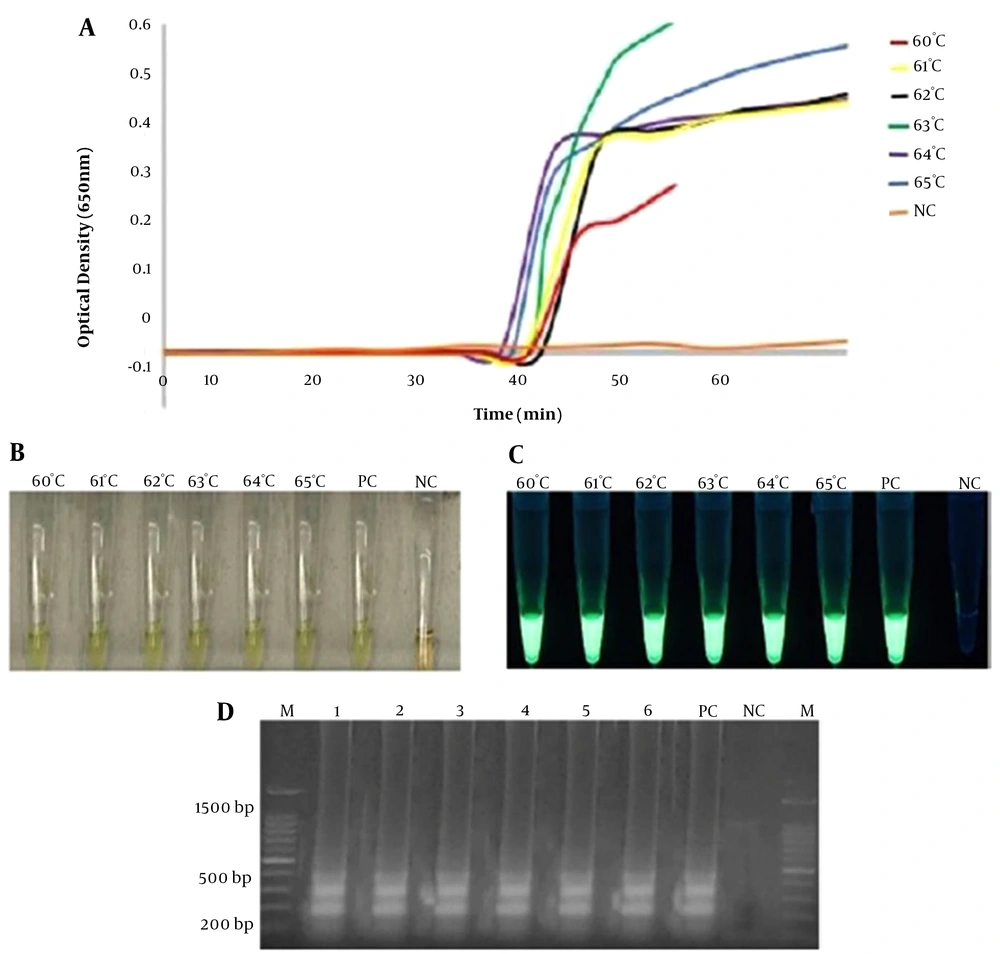
In the current study, RT-LAMP amplification products were monitored by observing the reaction or sigmoidal curve via a turbidimeter machine (LA-500, Eiken Co. Ltd, Japan) according to an established protocol (19). The results were analyzed using a web plot digitizer software (https://automeris.io/WebPlotDigitizer/) to generate data points, which then were subsequently plotted to the graph. Sigmoidal curves were observed for all different reaction temperatures (from 60 to 65°C) when the RT-LAMP assay was run with 100 ng of DNA template within 60 min in this study. The absorbance reading indicated that the highest level of amplification products was observed at 63°C as illustrated in Appendix 1A. This temperature was adopted for subsequent RT-LAMP assays in this study.
In addition, the amplification products in the reaction tubes were also directly observed by the naked eye and UV light. The presence of PCR products at different temperature conditions was observed by gel electrophoresis. The change of color from pale orange to green (positive reaction) was observed in reaction tubes in all temperature conditions, thus, confirming the formation of reaction as plotted sigmoidal curves via turbidimetry method. As for the evaluation of RT-LAMP assay at different reaction times, no curve was generated at 10 min of the optimization of reaction time, but RT-LAMP amplification was observed as early as 15, 30, 45, and 60 min (Figure 1A). Similar findings were also detected by other methods used in this study (Figure 1B-D).
Optimization of RT-LAMP assay according to reaction time temperature (a, b, c, d). Different line colors represent reaction times ranging from 10 min to 60 min in (a). The highest amplification was observed at 45 min and was chosen for the optimum time for the RT-LAMP assay. Other assessments were based on the visualization of color change (orange to green) and green fluorescence (calcein dye) in the reaction tubes by the naked eye (b) and UV light (c). Finally, the LAMP PCR products were visualized by 1.5% gel electrophoresis (d). A: 60 min, B: 45 min, C: 30 min, D: 15 min, E: 10 min M: a 100-bp ladder, PC: positive control, NC: negative control.
4.2. Specificity and Sensitivity of RT-LAMP Assay
A total of 11 strains were used to evaluate the specificity of the RT-LAMP assay for the detection of the speB gene. The results demonstrated the presence of sigmoidal curves for the DNA extracts of S. pyogenes (ATCC 19615) and clinical GAS isolate only. The two streptococci species, namely S. pneumoniae and S. agalactiae, along with seven other strains, generated no graphs throughout the entire 45 minutes of RT-LAMP assay. The findings were also consistent with those obtained using the calcein indicator (visualized by the naked eye and UV lamp), as well as by agarose gel electrophoresis as shown in Appendix 2.
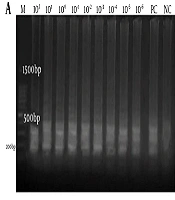
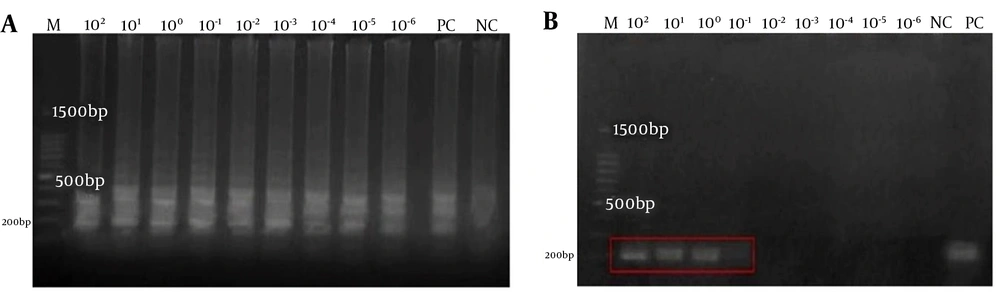
The sensitivity of RT-LAMP was compared with the conventional PCR. The sigmoidal curves were observed in all reaction tubes containing the 10-fold serial dilutions of total genomic DNA of S. pyogenes (ATCC 19615) ranging from 102 to 10 - 6 ng/µL compared to the conventional PCR assay (1 ng/µL only) (Appendix 3 in Supplementary File). Therefore, the RT-LAMP assay was 106-fold more sensitive than the conventional PCR assay in our study. Thus, the detection limit of the RT-LAMP for speB was 0.001 pg/µL compared to 1 ng/µL sensitivity of the conventional PCR (Figure 2). This is further correlated with the findings from visual observations (Appendix 3 in Supplementary File).
Sensitivity of RT-LAMP assay for the detection of the speB gene using a 10-fold serial dilution ranging 102-10-6 ng/µl in this study. Agarose gel electrophoresis of LAMP and conventional PCR products were compared in (a) and (b). M: a 100-bp ladder, PC: positive control, NC: negative control. The red box highlights the presence of PCR products at only 102, 101, and 100 ng/µl concentrations and the absence of PCR products at 10-1 ng/µl in the conventional PCR assay in (b).
4.3. Detection of speB by the PCR and RT-LAMP Assays
The speB gene was detected in all 43 (100%) S. pyogenes clinical isolates by conventional PCR assay in this study. Similarly, the sigmoidal curves indicating the presence of the speB gene were also observed in the RT-LAMP assay, as shown in Appendix 4. This is also visualized by the naked eye, UV light, and agarose gel electrophoresis (data not shown).
5. Discussion
Due to the global economic recession affecting many unfortunate countries, it is imperative to have a reliable, fast, and cost-effective method for the detection of GAS. Loop-mediated isothermal amplification is considered a novel approach for its cost-effectiveness, reliability, and high sensitivity of results (8). Loop-mediated isothermal amplification employs the auto cycling strand displacement DNA synthesis technique under isothermal conditions. It also utilizes a set of four to six specifically designed primers to hybridize six to eight different parts of the target DNA sequence, a feature that makes this detection technique specific to the desired gene (20). Several modifications to the LAMP method were invented to further enhance its novel features in microbial diagnostic fields (21-26).
In the present study, the combination of turbidimetry and LAMP methods could offer an accurate visual detection by observing the generation of a graph. The sigmoidal curve was generated due to the presence of magnesium ions (Mg2+) in the reaction buffer. During the LAMP reaction process, pyrophosphate ions are liberated to form a white magnesium phosphate precipitate that can be monitored by a turbidimetry method (14). It is known that false-positive results caused by various factors may limit the use of this highly sensitive LAMP method (27, 28). To overcome these limitations, the calcein dye was used in the present study, and a turbidimeter machine (LA-500, Eiken Co. Ltd, Japan) was utilized. The calcein dye was added to the LAMP reagents, and the color change (from orange to green) was observed in each reaction tube when the LAMP procedure was completed.
During PCR product amplification, the calcein/manganese ions (Mn2+) complex would be displaced by pyrophosphate ions, and the color would turn from orange to green (29). In addition, the turbidimeter machine (LA-500, Eiken Co. Ltd, Japan) in the present study utilized the concept of a closed system operation whereby the lid was opened once, the reaction tubes were tightly closed, and the cap was only opened once when the LAMP mixture was added into each reaction tube. Thus, this could reduce the contamination of amplification products via aerosols. The use of a colorimetric agent (calcein) in the reaction buffer would enhance the visibility of amplification products by the naked eye. This step aids the idea of using the LAMP method in resource-poor and developing countries (30).
The graph was generated as early as 15 min in the present study, which was also supported by the findings observed in other methods as shown in Figure 2. The time for speB gene detection in the present study was shorter than that of the colorimetry method used by Cao et al., which was 35 min (16). In their study, the LAMP method was applied, but the endpoint detection for the LAMP reaction was only based on the color change by naked eye observation (16). A longer duration of gene detection was also observed in a few studies. For instance, despite similar combination methods used, the detection time for the spy1258 gene of S. pyogenes was established at 24 min by Zhao et al. (31). In a study involving non-S. pyogenes isolates, the vanA gene of Enterococcus species was detected at 25 min after the completion of LAMP reaction. The vanA gene was visualized by gel electrophoresis as the end-point of the detection (32).
In the present study, the specificity of speB gene detection by the RT-LAMP assay is comparable with a conventional LAMP assay using the chromogenic method (color change) by Cao et al. (16). However, the sensitivity of the RT-LAMP assay in the present study is higher (0.001 pg/µL) than that of their study. They reported 100-fold sensitivity with the detection limit of 10 pg/µL in the conventional LAMP assay using the chromogenic method (color change) compared to conventional PCR (16). Another study on the RT-LAMP assay only yielded 100-fold sensitivity than the conventional PCR assay in detecting Shigella sp (19). This is not surprising as the LAMP method utilizes a highly specified set of primers that could bind to seven distinct regions on the target gene (speB); thus, the sequence selectivity and the high number of amplification products can be highly ensured (20).
The use of B. stearothermophilus DNA polymerase (Bst) in the RT-LAMP assay in the present study could also improve the amplification process, as it has a high degree of tolerance against inhibitory substances compared to other DNA polymerases such as Taq in the reaction mixture (33). In addition, the boiling method that was used for DNA extraction in the present study is also able to reduce chemical pollution, which could be normally found in the DNA extraction kits (16). It seems that the performance of the RT-LAMP assay varies according to the type of pathogen, the number of specific primers used LAMP assay method, type of visual observation, and the molecular methods used for comparison. Nonetheless, the increased sensitivity and the shorter duration of the target gene amplification (15 min) in the RT-LAMP assay in the present study could replace the conventional methods used for routine S. pyogenes diagnosis in clinical samples.
Apart from several advantages highlighted in the present study, there are also several limitations. First, the RT-LAMP assay was used to analyze only 43 S. pyogenes clinical isolates, thus, diagnosing a larger number of clinical isolates from different types of infections could establish a higher performance of RT-LAMP assay in the future. Second, the direct detection of S. pyogenes from clinical specimens by RT-LAMP assay was not performed in this study, but considering the robustness of the RT-LAMP mixture, it can be further investigated in future experiments. In addition, the preliminary finding presented here suggests that the speB gene RT-LAMP assay is rapid, easy to perform, and affordable with high sensitivity and specificity. Thus, it could be exploited for an ideal point-of-care method. It is with great hope that the rapid detection of S. pyogenes in clinical samples can substantially reduce nosocomial transmission and outbreaks, with the implementation of better management in the future.