1. Background
Breast cancer is the most common malignancy in females worldwide. Only in the United States, 279,100 new cancer cases and 42,690 related deaths were projected to occur in 2020 (1). Breast cancer can begin in different parts of the breast, but most of them start from milk ducts and are called ductal carcinoma, which is the most common type of breast cancer. Lobular carcinomas are the second most frequent histologic forms of this malignancy, which begin in milk-producing lobules (2, 3). Less-common types of breast cancer include tubular, mucinous, comedo, and medullary types (3). Tumor grades of I, II, and III are used to describe the rate of growth and spread of breast cancer cells (4). Breast cancer is a multifactorial event, and the role of viral infections in its etiology is largely proposed (5).
Approximately 12% of all human malignancies have a viral origin. Among them, DNA oncoviruses, including human papillomavirus (HPV) and Epstein-Barr virus (EBV), account for a notable proportion and impose a high disease burden that is becoming greater in the world (6, 7). Human papillomavirus, a member of the Papillomaviridae family, is a major etiological cause of cervical cancer. It possesses more than 150 genotypes, several types of them are more implied to elevate the tumorigenesis risk (High-Risk HPV; HR-HPV) (8). Epstein-Barr virus, a member of the Herpesviridae family, was discovered as the first human oncovirus detected in Burkitt lymphoma cells in 1964 (6). The involvement of EBV is well-documented in a variety of human cancers, including Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, and T cell lymphomas (6). Human papillomavirus and EBV infection can cooperatively participate in the pathogenesis of certain types of malignancies, such as breast carcinoma, due to their potential oncogenic effects on cells with an epithelial origin. Both HPV and EBV DNA oncoviruses are etiologically correlated with breast carcinoma development (9). Human papillomavirus and EBV DNA oncoviruses play a pivotal role in the pathogenesis of breast cancer. Accumulating evidence points out that these oncoviruses can co-exist in cancerous tissues of the breast and subsequently facilitate the initiation and progression of malignancies (10-12).
These viruses are capable of disrupting the diverse cellular machineries in the host. Inducing the inactivation of host tumor suppressor genes is a broad strategy that is exploited by oncoviruses in the development of cancers (13). One of the most important classes of these genes’ inactivation is related to tumor suppressor microRNAs (miRNAs). As known, miRNAs include a category of endogenous non-coding RNAs with ~ 20 - 25 nucleotides in length and the ability to regulate gene expression via base-pairing at 3′ UTRs (14). Based on specific tumor contexts and target base-pairing genes, miRNAs can act as tumor suppressors or oncogenes (15).
As a cluster of miRNAs, miR-143 and miR-145 are generated by two distinct co-transcribed miRNAs whose functions as tumor suppressors are intensively described (16). The suppressive effect of the miR-143/145 cluster on inhibiting the formation of epithelial tumors like breast cancer is clearly shown (17). In breast cancer cells, miR-143 and miR-145 synergistically repress cell proliferation and invasion; thereby, their downregulation arises malignancy-related hallmarks (18). Notably, the disruption of the miR-143/145 cluster has been observed in cancers associated with HPV and EBV human oncoviruses (19, 20).
Some infectious agents are well-described causes of chronic inflammation, which can derive carcinogenesis. Both HPV and EBV can establish persistent infection stimulating prolonged inflammation (21). These oncogenic viruses can upregulate various pro-inflammatory cytokines including IL-8 (22, 23). As a well-described pro-inflammatory cytokine, IL-8 is involved in breast tumors and with significant changes in serum level in these patients. Besides, IL-8 has a paracrine and/or autocrine tumor-promoting role in the manipulation of proliferation and survival of tumor cells (24). An elevated level of IL-8 motivates the expression of angiogenic growth factors and cell adhesion molecules, leading to angiogenesis and metastasis in breast cancer patients (24, 25). As known, CXR-1 and CXR-2 are the leading receptors of IL-8, which are expressed in all breast cancer cells, representing the promoted sensitivity of cancer cells to IL-8 compared to normal breast cells (26). Importantly, functional analysis systems have revealed the implication of tumor suppressor miRNAs such as miR-145 in the secretion level of pro-inflammatory IL-8 (27).
2. Objectives
We newly sought the possible effect of viral infections on IL-8 by targeting tumor suppressor miR-143 and miR-145 in the serum of breast cancer patients. This is a particular part of virus-induced breast carcinogenesis that has not been considered yet. Here, we investigated the correlation between the viral presence and expression level of tumor suppressor miR-143 and miR-145 and clinical outcomes and their further effects on serum IL-8 in breast cancer patients.
3. Methods
3.1. Study Population and Collection of Specimens
A total of 70 tissue samples (35 cancerous tissues and 35 adjacent non-cancerous tissues) were used in the investigation collected from 35 women who had been diagnosed with breast carcinoma and had not undergone previous chemotherapy. Specimens were obtained from the primary tumors of subjects who underwent biopsies at different hospitals in Ahvaz province, Iran, from April 2020 to October 2020. The new cases with confirmed pathological evidence were included, and the patients who had a cancer history or metastatic cancer or received neoadjuvant chemotherapy were excluded.
In all women diagnosed with breast cancer, a breast examination was performed by an experienced surgeon. In addition, tissue samples were evaluated for the histopathologic assessment of breast cancer according to the WHO criteria by two experienced pathologists to ensure the correct diagnosis. All tissue specimens were immediately frozen in liquid nitrogen after biopsy and stored at -80°C for subsequent research. Also, all patients signed an informed consent form concerning the participation in the study. Before the needle biopsy, for all the enrolled women (n = 35), serum samples were collected and stored at -80°C until analysis. We also collected 35 serum samples from healthy normal individuals/volunteers (without a history of malignancy and chronic inflammatory diseases) as a serum control group to determine the difference in cytokine levels between patients and normal subjects.
3.2. Genomic DNA Preparation, Polymerase Chain Reaction, and Genotyping
The DNA extraction was carried out using QIAamp DNA Mini Kit (Cat No./ID: 51304) based on the manufacturer's protocols. The amplification of the human β-globin gene was performed using PCO3/PCO4 primers (Table 1) to evaluate the quality of the extracted material, and positive samples underwent further investigations. Specimens were screened for the presence of HPV utilizing the nested PCR method with primers specific to the L1 region of the HPV genome (Table 1). For both rounds, PCR conditions included 1X PCR buffer, 300 - 400 ng of template DNA, 1 mM MgCl2, 100 mM deoxynucleotide triphosphates (dNTPs), 10 pmol of each primer, and 0.5 U of Taq polymerase in a total volume of 25 µL. The first-round PCR thermal profile was 95°C for 7 min, followed by 40 cycles of 94°C for 45 s, 50°C for 45 s, 72°C for 45 s, and a final extension for 7 min at 72°C. The second round started with 95°C for 5 min, followed by 40 cycles of 94°C for 30 s, 50°C for 30 s, 72°C for 30 s, and a final extension for 7 min at 72°C.
| Locus | Oligonucleotide Sequence | ORF | Fragment Size | Reference |
|---|---|---|---|---|
| β-globin | β-globin | 110 | (28) | |
| PCO3 | ACACAACTGTGTTCACTAGC | |||
| PCO4 | CAACTTCATCCACGTTCACC | |||
| EBV | EBNA1 | 609 | (29) | |
| F1 | GTAGAAGGCCATTTTTCCAC | |||
| R1 | CTCCATCGTCAAAGCTGCA | |||
| EBV | EBNA1 | 308 | - | |
| F2 | AGATGACCCAGGAGAAGGCCCAAGC | |||
| R2 | CAAAGGGGAGACGACTCAATGGTGT | |||
| HPV | L1 | 450 | (30) | |
| F1 | CGTCCMARRGGAWACTGATC | |||
| R1 | GCMCAGGGWCATAAYAATGG | |||
| HPV | L1 | 150 | - | |
| F2 | TTTGTTACTGTGGTAGATACTAC | |||
| R2 | GAAAAATAAACTGTAAATCATATTC |
For EBV detection, the EBNA-1 coding region was amplified (bp) by nested PCR, using specific primers (Table 1). Each PCR was performed in 25 µL of a reaction mixture containing 25 pmol of each of outer and inner primers, 100 μM dNTPs, 1X PCR buffer, 1.5 mM MgCl2, one unit of Taq polymerase enzyme (Amplicon, USA), and 300 ng of extracted/product DNA. The PCR cycling protocol included 10 min at 94°C, followed by 45 s at 94°C, 45 s at 53°C, and one minute at 72°C for 35 cycles in the first PCR, and 5 min at 94°C, 45 s at 94°C, 45s at 63°C, and one minute at 72°C for 35 cycles in the second PCR. In the next step, PCR products were excised from an agarose gel (Figures 1 - 3) and purified. Subsequently, they underwent direct sequencing using ABI PRISM 3100 (Applied Biosystems) to confirm the viral presence and determine the HPV genotype.
3.3. RNA Isolation and Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
In this study, miRNAs were extracted from cancerous tissues and the matched normal tissue specimens, using the RNA Minipreps Kit (Bio Basic, Inc.) according to the kit instruction. The concentration and purity of RNA were measured with NanoDrop™ via N-D 2000 (Thermo Scientific, USA). In this step, 1 µg of extracted RNA template was reverse-transcribed to cDNA in a final volume of 20 µL, employing the BON-Stem miR cDNA synthesis (Cat No. BN-0011.17.2, Iran) based on the manufacturer’s protocol. To evaluate the gene expression of the miR-143/145 cluster, RT-qPCR was carried out with the BON-miR QPCR Kit (Cat No. BON209002, Iran) in a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, USA). The PCR reaction mixture was in a final volume of 13 μL containing 1 μL of cDNA, 0.5 mM of each primer, and 2 × miRNA QPCR master mix Passive reference dye I (ROX) (Cat No. BN-0011.17.3, Iran). The amplification conditions were 5 min at 95°C, 40 cycles of 5 s at 95°C, and 30 s at 58°C, followed by a melting curve cycle. The reactions were normalized to the expression of endogenous control SNORD47. Each test was performed in duplicate. The 2-ΔΔCt formula was used to determine the expression levels of miR-143 and miR-145.
3.4. Cytokine Assessment
All sera taken from patients (n = 35) and normal controls (n = 35) underwent testing for circulating IL-8 levels by the Human IL-8 ELISA® Kit (Karmania Pars Gene, Iran), based on the manufacturer’s instructions with a sensitivity of 3.0 pg/mL. The IL-8 level of each serum specimen was examined two times and an average was reported.
3.5. Statistical Analysis
The normality of data was tested using the Kolmogorov–Smirnov test for continuous variables. The Mann-Whitney U test was used to assess the differences in miR-143, miR-145, and cytokine levels between patients and controls. The chi-square test was used to assess the association of the groups with viruses, as well as with clinicopathological factors of patients. All data were evaluated using the Statistical Package for the Social Sciences (SPSS), version 22 (IBM), and Graphpad Prism, version 8.0.2 (Graphpad Software, Inc.). The P-values < 0.05 (*), < 0.01 (**), < 0.001(***), and < 0.0001(****) were considered statistically significant.
4. Results
4.1. Clinical Parameters and Screening for Virus Presence
A total of 70 fresh frozen breast carcinoma tissue biopsies from 35 patients were included in this research. The patients’ age ranged from 24 to 68 years in the cancerous group (mean 47.4 ± 12.7 y) and 27 to 64 in healthy individuals (mean 45.3 ± 14.7 y). The clinical data and presence of viral infection are summarized in Table 2. The screening of the viral genome for HPV and EBV showed that 14.2% (10/70) and 7% (5/70) were positive, respectively (Figures 2 and 3, Table 3). The detection of HPV DNA was significantly different between cancerous tissues (cases) and adjacent non-cancerous tissues (controls), while no significant difference was observed for EBV DNA (Table 3). Although two samples possessed a co-infection of HPV and EBV, its correlation with the case and control groups was not significant (Table 3). Human papillomavirus genotyping indicated HPV 16 (n = 5), HPV 73 (n = 2), HPV 53 (n = 1), and HPV 31 (n = 1) in the case group and HPV 16 (n = 1) in the control group.
| Parameter | Total, n = 35 | HPV+, n = 10 | HPV-, n = 25 | EBV+, n = 5 | EBV-, n = 30 | HPV/EBV, n = 2 |
|---|---|---|---|---|---|---|
| Age (y) | 47.4 ± 12.7 | 51.1 ± 13.3 | 46.8 ± 12.5 | 55 ± 10.2 | 46.8 ± 12.6 | 48.5 ± 13.4 |
| Type of cancer | ||||||
| Ductal | 31 (88.5) | 7 (22.5) | 24 (77.4) | 4 (12.9) | 27 (87) | 2 (6.4) |
| Lobular | 2 (5.7) | 1 (50) | 1 (50) | 0 | 2 (100) | 0 |
| Medullary | 1 (2.9) | 1 (100) | 0 | 0 | 1 (100) | 0 |
| Tubular | 1 (2.9) | 0 | 1 (100) | 0 | 1 (100) | 0 |
| Grade | ||||||
| I | 15 (42.9) | 2 (13.3) | 13 (86.6) | 2 (13.3) | 13 (86.6) | 0 |
| II | 8 (22.9) | 3 (37.5) | 5 (62.5) | 1 (12.5) | 7 (87.5) | 1 (12.5) |
| III | 12 (34.3) | 4 (33.3) | 8 (66.6) | 1 (8.3) | 11 (91.6) | 1 (8.3) |
| Metastasis | 6 (100) | 5 (83.3) ** | 1 (16.6) | 1 (16.6) | 5 (83.3) | 1 (16.6) |
a Values are expressed as No. (%) unless otherwise expressed.
b The P-value < 0.01 (**) were considered statistically significant.
| Normal/Tumor Tissue | HPV and EBV Detection | ||
|---|---|---|---|
| HPV | EBV | HPV/EBV Co-infection | |
| Normal | 1 (2.9) | 1 (2.9) | 0 |
| Tumor | 9 (25.7) | 4 (11.4) | 2 (5.8) |
| P-Value | 0.006 ** | 0.164 | 0.151 |
a Values are expressed as No. (%) unless otherwise expressed.
b The P-value < 0.01 (**) were considered statistically significant.
4.2. Gene Expression of miR-143/145 Cluster
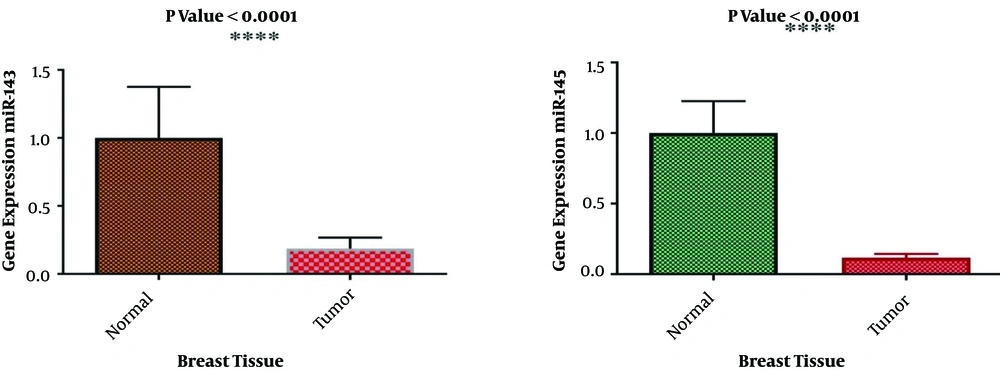
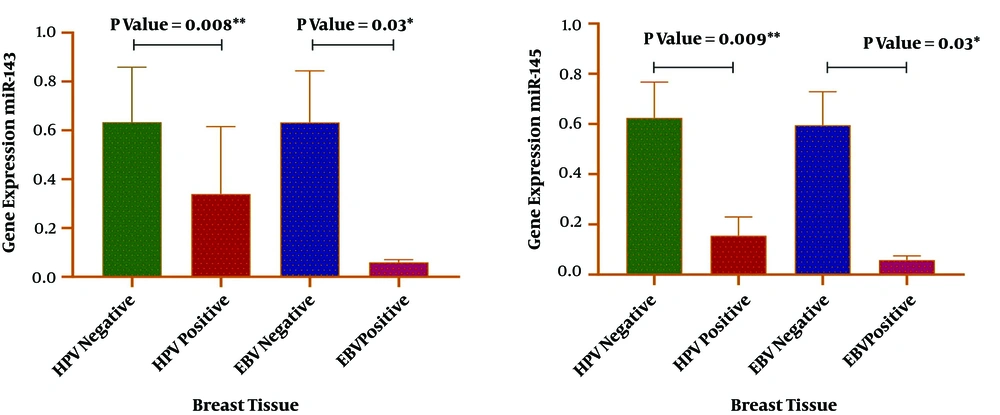
The expression levels of miR-143 and miR-145 were determined via RT-qPCR in cancerous and adjacent non-cancerous tissues of breast cancer patients. The relative expression of miR-143 and miR-145 was normalized by SNORD47. In comparison with adjacent non-cancerous tissues, the expression levels of both miR-143 and miR-145 were reduced in the breast cancer group, and the difference was statistically significant (P < 0.0001). The results of miR-143/145 expression in the tumor and normal groups are shown in Figure 4. The analysis showed a direct association between HPV and EBV positivity and a reduced expression level of miR-143 and miR-145 (Figure 5). In addition, there was a correlation between the reduced expression of the miR-143/145 cluster and metastasis according to HPV positivity (P = 0.003).
4.3. Serum Interleukin-8
Interleukin-8 concentrations were determined by enzyme-linked immunosorbent assays (ELISA) in serum specimens. Serum concentrations of IL-8 were almost similar between patients and controls (P = 0.296) (Table 4). We found no statistically significant correlation between patients who were infected with HPV/EBV and non-infected patients according to the concentration of IL-8 in serum (Table 4). Additionally, no correlation was found between the IL-8 serum level and clinicopathological data.
| Group | Mean ± SD | P-Value |
|---|---|---|
| Control | 15.4 ± 7.6 | 0.14 |
| Case | 18.7 ± 12.2 | |
| HPV+ | 24.2 ± 21.1 | 0.788 |
| HPV- | 16.5 ± 5.2 | |
| EBV+ | 22.5 ± 18.6 | 0.766 |
| EBV- | 18.1 ± 11.2 |
5. Discussion
Human papillomavirus and EBV DNA oncoviruses account for nearly two-thirds of all virus-associated malignancies (31). Human papillomavirus and EBV can co-exist and cooperate in human cancers, especially breast cancer (32). The prevalence of HPV in breast cancer varies widely from 1.6 to 86.2% among countries around the world (33). This rate in Iran has been reported in various studies from 6.7 to 40.5%, which is considered to be 23.7% on average (34). However, the prevalence of EBV in different countries of the world among women with breast cancer is reported from 0 to 78.1%, and 26.3% on average. It has the highest prevalence in Europe and the lowest in North America. In various studies in Iran, different prevalence rates have been reported from 0 to 35.3% (35).
Carcinogenic properties of these DNA oncoviruses allow them to drive cell transformation and tumor development. Alterations in host cell miRNA expression can be done by HPV and EBV as a pivotal mechanism involved in virus-mediated oncogenesis (36). The current research was undertaken to investigate the correlation between the presence of oncoviruses, HPV and EBV, and probable dysregulation of tumor suppressor miRNAs, miR-143 and miR-145, in breast cancer patients and evaluate their potential link to the clinical outcome. In the first step, we explored the prevalence of HPV and EBV in breast cancer tissue and corresponding adjacent non-cancerous tissues using nested-PCR. Human papillomavirus was significantly different between the case and control groups (Table 3).
There is a range of discrepancies in the literature concerning the HPV prevalence in breast cancer patients. In line with our data, investigations involving cases and controls indicate a higher frequency of HPV in tumor tissues (37-41), while there are some studies that reported contradictory data (42, 43). The distribution of the HPV genotype in this research indicated that all detected types belonged to high-risk HPV. Human papillomavirus 16 was the most abundant type present in the patients. This HPV type has been frequently detected as the most prevalent type in other studies of breast cancer (39, 44).
For EBV, the screening of the viral genome did not show any correlation with the case and control groups (P = 0.164) (Table 3). The rate of EBV infection in breast cancer patients was not unexpected, as the virus has been detected in many studies to be absent or be at a low rate at this site (45-47). However, we did not observe any significant association between breast cancer and EBV presence. The more frequency rate of EBV and HPV in cancerous tissue may represent the increasing role of EBV in the development of breast cancer. For instance, an investigation conducted by Makielski et al. in oral cancer (48) proposed that infection with HPV may raise the capacity of epithelial cells to assist the EBV life cycle that could, in turn, elevate EBV-related pathogenesis in malignancy. In this regard, another study demonstrated that HPV/EBV coinfection enhanced EBV persistency via either latency or increased virus replication, as well as by elevating HPV oncogene expression (49). Therefore, EBV co-presence with HPV infection, even at low levels, might affect the susceptibility of breast epithelial cells to carcinogenesis.
Our data further indicated that less expression of these miRNAs was associated with a positive status of HPV in patients (Figure 5). Our results are consistent with data in the literature, indicating that miR-143 and miR-145 are largely dysregulated in HPV-associated cancers. Wang et al. demonstrated that the downregulation of miR-143/145 is a key factor for tumor growth in HPV-related cervical carcinogenesis (19). Baffa et al. also mentioned a reduced expression of miR-143/145 in HPV-related head and neck cancers (50). Reports imply the role of viral oncoproteins as a cause of suppressing the miRNAs. For example, it has been observed that the inhibition of HPV oncoproteins (E6 and E7) expression may trigger miR-143 upregulation in infected cells (51).
Intriguingly, we also realized a correlation between the reduced expression of the miR-143/145 cluster and metastasis according to HPV positivity (P = 0.003). The available data point out the capacity of HPV for triggering metastasis and inducing more aggressive phenotypes in breast carcinoma (31). Furthermore, the evidence demonstrated that miR-143/145 expression was negatively related to lymph node metastasis, and patients with lower levels of miR-143/145 expression may be subject to poor prognosis malignancies (52, 53). Collectively, our findings may present a novel mechanism by which HPV provokes breast cancer metastasis.
Therefore, considering the confirmed relationship between HPV and decreased miR-143/145 expressions in various malignancies and due to the close association between miR-143/145 downregulation and breast cancer, the findings of the current study may suggest the participation of HPV in the pathogenesis of breast carcinoma and adverse clinical outcomes such as the promotion of metastasis. Further analysis also showed less expression of miR-143/145 in the EBV-positive group as compared to the EBV-negative group (Figure 5). Very few reports have highlighted the correlation between EBV infection and the expression status of miR-143/145 (54). A study conducted in 2015 by Bazot et al. showed EBV capacity in silencing the expression of the miR-143/145 cluster in infected cells (20). They proposed EBV EBNA3A and EBNA3C protein implications in this process. Although herein, a significant result was gained in this respect, further studies seem to be needed to precisely evaluate the impact of EBV on these tumor suppressor miRNAs in breast carcinoma development.
In the final step, we speculated whether viral infection, especially HPV, can affect IL-8 production systematically via the disruption of two tumor suppressor miRNAs (miR-143 and miR-145). The elevation in the IL-8 serum level is suggested to be related to the development and progression of breast carcinogenesis (24). For this purpose, we obtained serum specimens from all patients in the current investigation and healthy controls. The comparative analysis based on the IL-8 serum concentration within different groups (Table 4) did not show any significant difference. Although some data have shown the promotion of serum levels of IL-8 upon viral persistent infections such as HPV and EBV (55, 56), studies remark the differences in the biology of viral infections in different types of cancers and the sensitivity of testing methods.
5.1. Conclusions
Our study may present strong data for the engagement of DNA oncoviruses, including HPV and EBV, in the promotion of breast carcinoma and metastasis via downregulating miR-143/145. Collectively, the results may suggest the use of viral vaccination for prevention or prescription of antiviral regimes in combination with other breast cancer therapies for infected people and application of miR-143/145 as a useful predictive biomarker for breast cancer outcome. However, further studies seem to be necessary.