1. Background
Fluoroquinolones are broad-spectrum antibiotics against Gram-positive and Gram-negative bacteria. In these bacteria, DNA topoisomerase IV and DNA gyrase are the main targets of fluoroquinolones, such as ciprofloxacin (1, 2). Ciprofloxacin inhibits the progression of DNA replication and transcription, producing DNA breaks and R-loops (RNA-DNA hybrids) in Gram-negative bacteria, such as Escherichia coli (3, 4). These changes induce the SOS response by promoting the RecA-dependent self-cleavage of LexA, as the repressor of SOS genes, including recA, dinB, and umuCD; the last two genes encode SOS DNA polymerases, namely DNA polymerase IV (pol IV) and DNA polymerase V (pol V). Pol V, as a complex enzyme, consists of UmuC and UmuD’2 dimers. These proteins are encoded by umuCD operon and activated after self-cleavage and dimerization of UmuD (4, 5).
SOS DNA polymerases participate in error-prone mutagenic DNA repair processes, such as translation synthesis (TLS), to restart replication forks that are stalled at damage sites or to repair gaps that have left the replication forks behind. Pol IV acts before pol V in the replisome region, and by increasing the concentrations of UmuD2 and UmuD’2, the access of pol IV to the replisome regions is prevented (6). Moreover, pol IV participates in error-prone double-strand break repair and transcription-coupled TLS. Therefore, DinB interacts with RecA and NusA during strand exchange activities and gap repair, respectively (7). The gap produced in the transcribed strand, opposite to a lesion in the non-transcribed strand, prevents the progression of the transcription complex. A component of this complex, which is a modulator of RNA polymerase activity (NusA), recruits DinB to fill the gap (8).
Activation of DNA pol IV and pol V confers resistance to ciprofloxacin through accumulation of several mutations in the target genes, encoding the subunits of DNA gyrase and topoisomerase IV, as well as regulatory genes (marR and acrR), inhibiting the overexpression of the AcrAB-TolC efflux pump (4, 9). The overexpression of this pump causes multi-drug resistance to structurally irrelevant antibiotics, such as tetracycline and rifampin. Rifampin (also called rifampicin), a derivative of rifamycin, is one of the most potent and broad-spectrum antibiotics, which binds to RNA polymerases (RNAP) and inhibits the transcription process (10, 11). The acquired mutation in genes, encoding the subunits of RNAP, especially rpoB gene that encodes the β subunit of enzyme, confers resistance to rifampin. These mutations in the rifampin resistance-determining region of rpoB, which mainly consist of cluster I and cluster II amino acid residues (located deep within the DNA-RNA channel of RNAP), reduce the affinity of enzymes for rifampin and affect the elongation and termination steps of transcription.
The rifampin naphthyl moiety contacts βresidues, including Val146, Leu511, Ser513, Arg529, Ser531, Leu533, Gly534, Asn568, and Ile572. On the other hand, the antibiotic ansa moiety contacts other βresidues, including Arg143, Glu510, Leu511, Leu512, Phe514, Asp516, Thr525, His526, Pro564, and Pro564 (11). Mutations in these sites are associated with certain phenotypes, such as slow growth, cold sensitivity, and stringent response mimicry phenotypes. Nonetheless, the last phenotype may not confer rifampicin resistance. Therefore, depending on the site of mutation, the minimum inhibitory concentration (MIC) for rifampin varies.
Substitutions at three sites, that is, Asp516, His526, and Ser531, cause high levels of resistance to rifampin and are associated with little or no loss of fitness (11). These mutations have been frequently detected in the clinical isolates of Rif-resistant (Rifr) bacteria. For example, the Ser531-to-Leu substitution has been frequently detected in the clinical isolates of Mycobacterium tuberculosis. Moreover, other mutations in rpoA, rpoB, and rpoC genes, encoding α (RpoA), β(RpoB), and β’ (RpoC) subunits of RNAP, respectively, have been also found in clinical isolates. Most of the secondary mutations in RpoC have been identified in the region interacting with RpoA (12). Important regions in the β subunit of RNAP remain conserved in bacteria (13).
In Gram-negative bacteria, umuC and umuD are mostly located in the same operon and are overexpressed differently late in SOS response (level of UmuD is higher than UmuC). However, levels of UmuD2 and UmuC are kept to a minimum through their proteolysis by the Lon protease in undamaged cells. In damaged cells, UmuD’ in UmuD/D’heterodimer is rapidly degraded by the ClpXP protease (14, 15). On the other hand, umuD is not found in Gram-positive bacteria, such as Streptococcus and Staphylococcus species. Therefore, unlike E. coli, DNA pol V is not the main mutagenic polymerase in these bacteria following exposure to ciprofloxacin. Ciprofloxacin induces mutagenesis in Streptococcus species through umuC-dependent or UmuC-independent mechanisms (5). The independent mechanism of S. pneumonia and S. uberis depends on acquiring mutations in the rifampicin resistance-determining region of rpoB gene in wild type strain and umuC (ΔumuC) mutant (16). It is not clear whether ciprofloxacin treatment of E. coli mutants lacking active umuC can cause rifampin-resistant mutations.
2. Objectives
We aimed to investigate whether the formation of rifampin-resistant mutations in E. coli umuC mutants after exposure to ciprofloxacin would compensate for the mutagenesis defect.
3. Methods
3.1. Bacterial Strains and Growth
The bacterial strains used are listed in Table 1. Among them Escherichia coli BW25113 and a knockout mutant JW11731 (BW25113 ΔumuC:Kanr) were obtained from the Keio collection (E. coli Genetic Stock Collection (CGSC), USA) (17). Clones with higher levels of resistance to ciprofloxacin were generated from JW11731 in presence of increasing amounts of ciprofloxacin (Sigma, USA) up to 2 µg/mL ciprofloxacin (18).
| Strain/Mutant | Genotype | MIC, µg/mL | Source/Reference | |
|---|---|---|---|---|
| Cip | Rif | |||
| MG1655 | Wild type | 0.035 | 0. 4 | A gift from Prof. RG Lloyd |
| BW25113 | Parent strain | 0.04 | 0.4 | Keio collection |
| JW11731 | BW25113 ΔumuC:Kanr | 0.04 | 0.4 | Keio collection |
| M2 | gyrA (Ser83 → Leu) marOR (20 bp duplication) acrAB overexpression | 100 | 100 | Pourahmad Jaktaji and Psand 2016 |
| MN1 | JW11731 ΔumuC:Kanr | 0.3 | 1 | This work |
| MN2 | JW11731 ΔumuC:Kanr | 2 | 16 | This work |
| MN3 | JW11731 ΔumuC:Kanr | 2 | 16 | This work |
| MN4 | JW11731 ΔumuC:Kanr | 2 | 30 | This work |
| MN5 | JW11731 ΔumuC:Kanr | 2 | 50 | This work |
Abbreviations: Cip, ciprofloxacin; MIC, minimum inhibitory concentration; Rif, rifampin.
3.2. Minimum Inhibitory Concentration
Minimum inhibitory concentration of ciprofloxacin and rifampin (Merck, Germany) for E. coli strains and mutants were determined by the broth dilution method in accordance to Clinical and Laboratory Standards Institute (CLSI) guidelines (19). Briefly, Luria-Bertani (LB) inoculated with 5 × 107 - 5 × 108 CFU/mL of bacteria was used. Depending on bacterial genetic background, serial dilutions of ciprofloxacin (from 5 ng/mL to 2 µg/mL) and rifampin (from 10 ng/ml to 60 µg/mL), were added to cultures and the cultures were incubated for 16 - 24 h at 37°C. The experiments were conducted three times.
3.3. PCR Amplification and DNA Sequencing
The rifampin resistance-determining region of rpoB gene (containing cluster I and II of the β subunit of RNA polymerase) was amplified by colony PCR method. The following primers were used for rpoB: forward primer, 5’-TGATCAACGCCAAGCC-3’ and reverse primer, 5’-TACAACACCGTCGGTC-3’. The amounts of PCR ingredients were: 2 µL of DNA, 5 µL of PCR buffer, 015 µL of Taq polymerase (genfanavaran, Iran), 0.5 µL of each primer, 0.5 µl of MgCl2 and 0.5 of dNTP. The final volume of reaction mixture was reached to 25 µL with sterile distilled water. The thermal condition was: pretreatment 95°C for 3 min, amplification steps, 94°C for 45 s, 52°C for 30 s and 72°C for 1 min, for 30 cycles and final extension 72°C for 4 min. The sizes of amplified segments should be 323 base pairs. Then PCR products were visualized by gel electrophoresis, then both strands of PCR products were sequenced by DNA sequencing with the forward and reverse primers and the nucleotide sequence of them was compared to that of MG1655 strain, which is retrieved from NCBI. The sequence of cluster I and II of rpoB gene was presented previously (10, 11).
3.4. Ciprofloxacin-induced Mutagenesis
Ciprofloxacin-induced rifampin resistance assays was conducted as described previously (20). Three independent exponentially growing cultures of MG1655, umuC clones (carrying the umuC mutation) and M2 in LB without and with ciprofloxacin (0.1 × MIC) incubated for 4 hours at 37°C with shaking (250 rpm). The bacterial cells were collected by centrifugation (for 10 min at 6000 rpm) in centrifuge (Sigma, Germany). The pellet was resuspended in fresh LB broth and incubated overnight at the same conditions described above. Viable cells were determined after preparing appropriate dilutions and plating on LB agar with and without rifampin (20 µg/mL); and incubated overnight at 37°C. The numbers of colonies were counted and the frequency of mutations, the ratio of resistant cells to viable cells, was calculated.
3.5. RNA Sample Preparation
Fresh cultures in LB broth containing sub-inhibitory concentration of ciprofloxacin were prepared. Cultures were grown to a mid-logarithmic phase (OD600 of 0.5 - 0.6). Appropriate volumes from each of cultures were mixed with 2 volumes of RNA protect reagent (Qiagen, Germany) and were centrifuged. Cell pellets were used for RNA extraction using an RNeasy Mini Kit (Qiagen, Germany). The quality and concentration of RNA was determined using the Ultrospec 1100 spectrophotometer (Japan).
3.6. Real Time PCR (qRT-PCR)
RNase-free DNase (Fermentas, USA) enzyme was used to eliminate DNA from RNA samples based on the manufacturer’s guideline. Then RNA samples were repurified using an RNeasy Mini kit (Qiagen, Germany). To detect possible DNA contamination RNA samples were amplified by PCR and visualized by gel electrophoresis. The cleaned RNA samples were used for cDNA synthesis. Reverse transcription was performed using the RevertAid Reverse Transcriptase Kit (Thermo scientific, USA), random hexamer and purified total RNA (500 ng). To test the quality of cDNAs, they were amplified by PCR reaction with specific primers.
Primers for recA, umuD, and gapA are described previously (18). Primers for dinB were as follows: Forward primer, 5’-CTCGCGCTACACCTCG-3’ and reverse primer, 5’-ACGCCGTCA GTTGCAG-3’. To determine gene expression by real time PCR, Rotor Gene 6000 thermocycler (Corbett Research, Australia) and SYBR Green kit (Takara, Japan) were used. Thermal cycling conditions were described previously unless for annealing temperature, which differs from one gene to another (18). Melting curve analysis at 60°C - 95°C with continuous fluorescence reading was conducted. Relative gene expression was calculated based on Pfaffl method (21). All data on gene expressions are the average of duplicate analyses.
3.7. Statistical Analysis
Statistically significant differences in gene expression were determined by Student’s t-test (two paired samples, with two tailed distribution), using SPSS version 16 software.
4. Results
4.1. Generation of Ciprofloxacin Resistant Clone
Clones, MN1-MN5 with higher resistance to ciprofloxacin were generated from JW11731 in the presence of increasing amounts of ciprofloxacin (Table 1). Exposure to more than 2 µg/mL ciprofloxacin, did not result in generating higher resistant clones.
4.2. Susceptibility of Strains and Mutants to Ciprofloxacin and Rifampin
Minimum inhibitory concentration of ciprofloxacin and rifampin for E. coli strains and mutants were presented in Table 1. MN5 showed about 50-fold higher MIC than the parental strain (JW11731) to ciprofloxacin and rifampin. Based on this result, MN5 showed intermediate level of resistance to ciprofloxacin and high level of resistance to rifampin based on previously reported data (22). However, M2 exhibited approximately 100-fold increases in MIC for rifampin as compared with wild type strain MG1655, due to overactivation of AcrAB-TolC efflux pump. As expected, MG1655 and M2 indicated low and high level of resistance to rifampin. Moreover, MN1 and MN2-MN4 showed low and high levels of resistance to rifampin, respectively. Therefore, it was possible to produce clones with high levels of resistance to rifampin from rifampin-sensitive umuC mutant strain.
4.3. PCR Amplification and DNA Sequencing
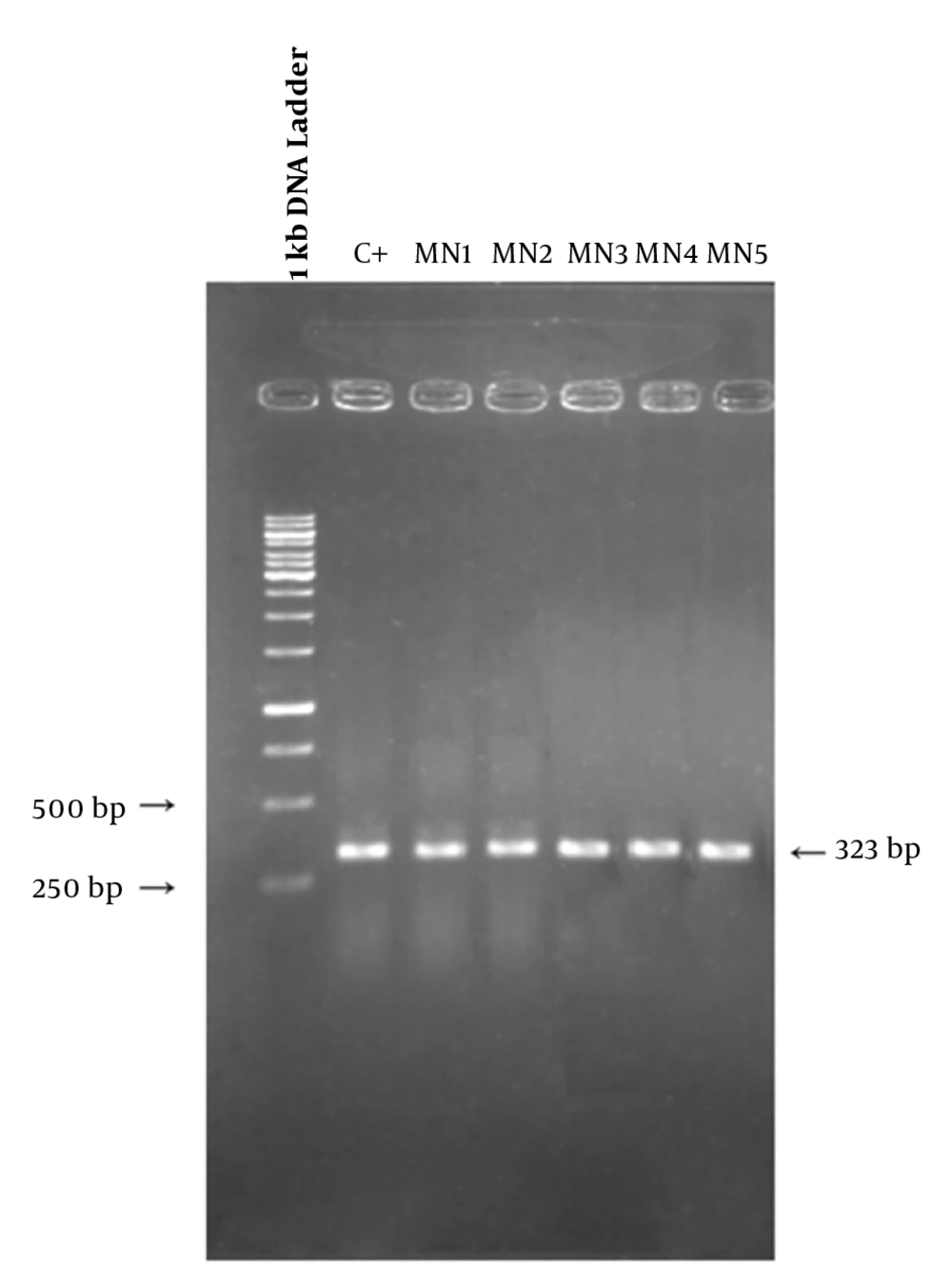
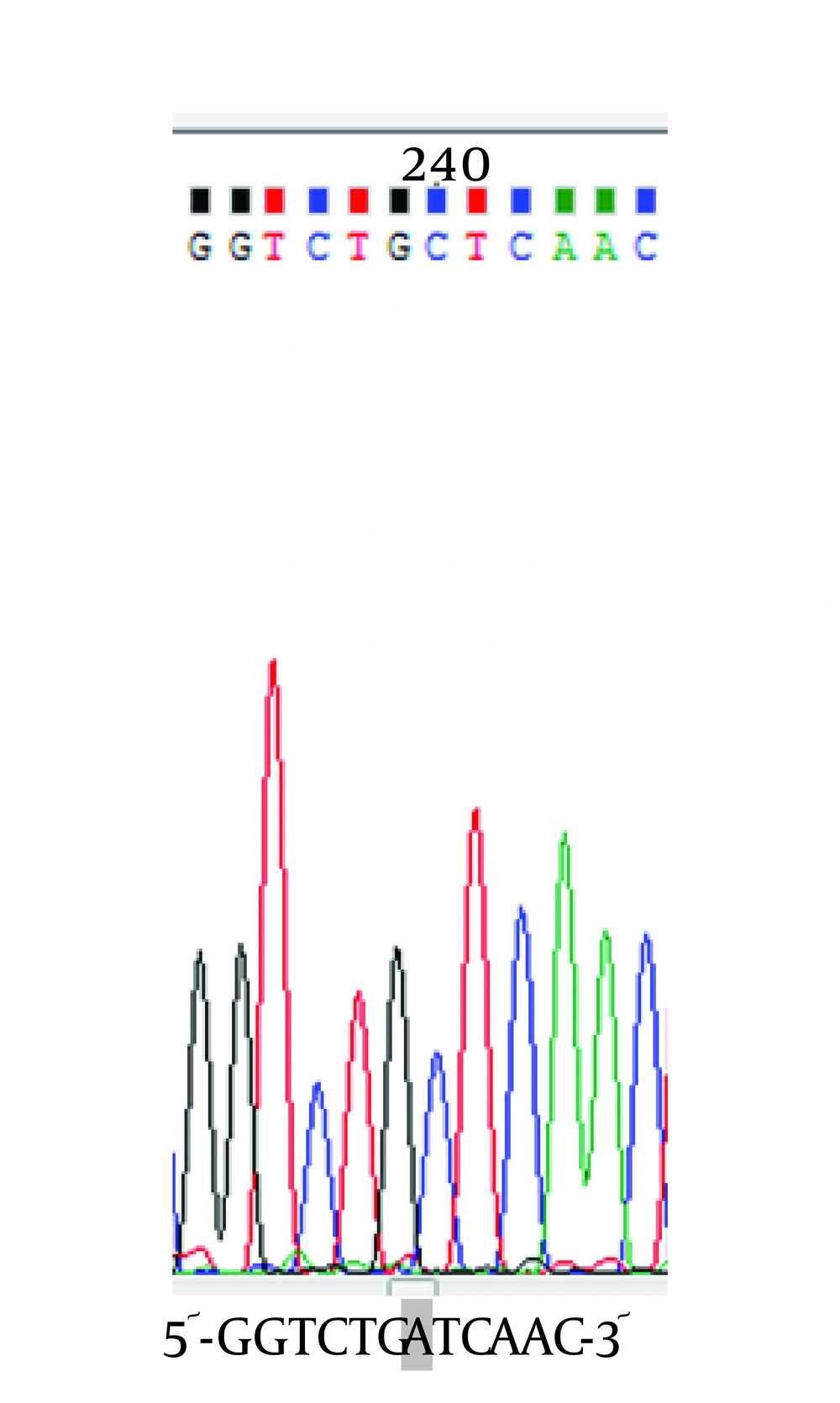
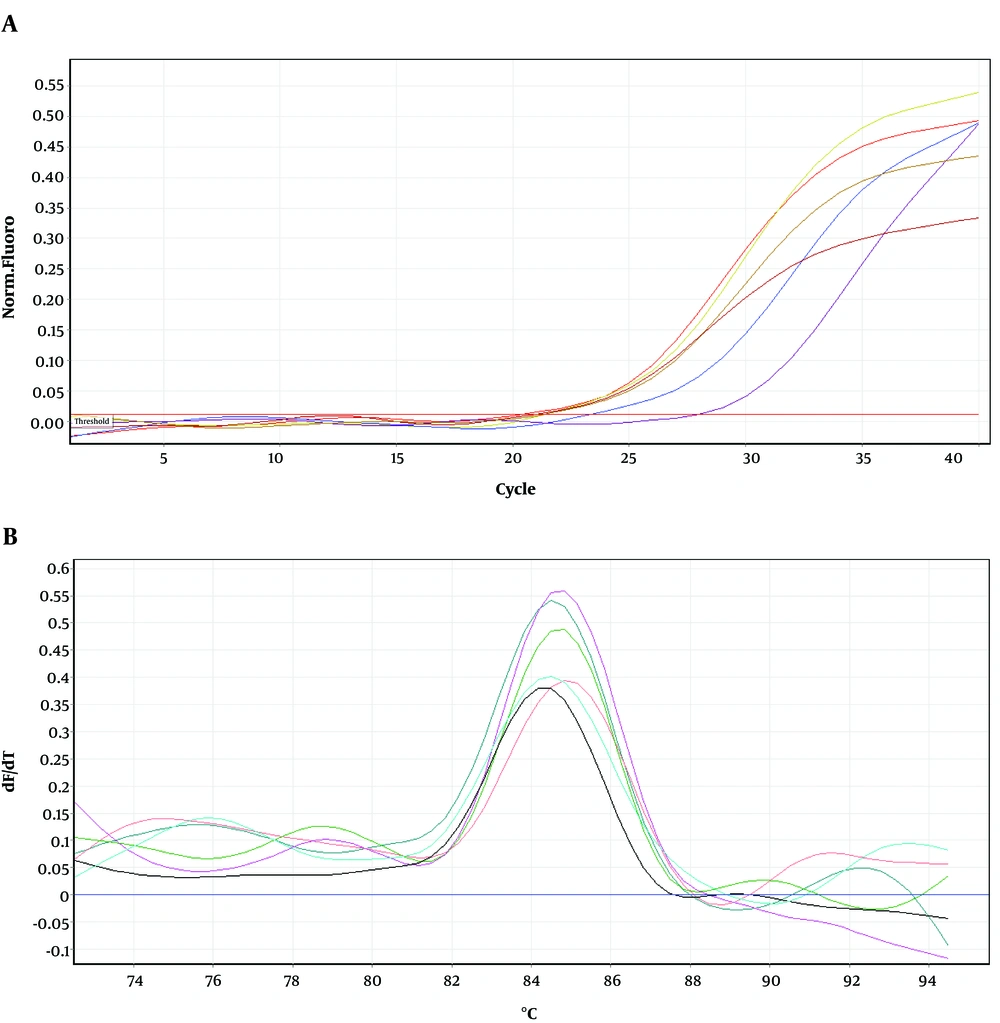
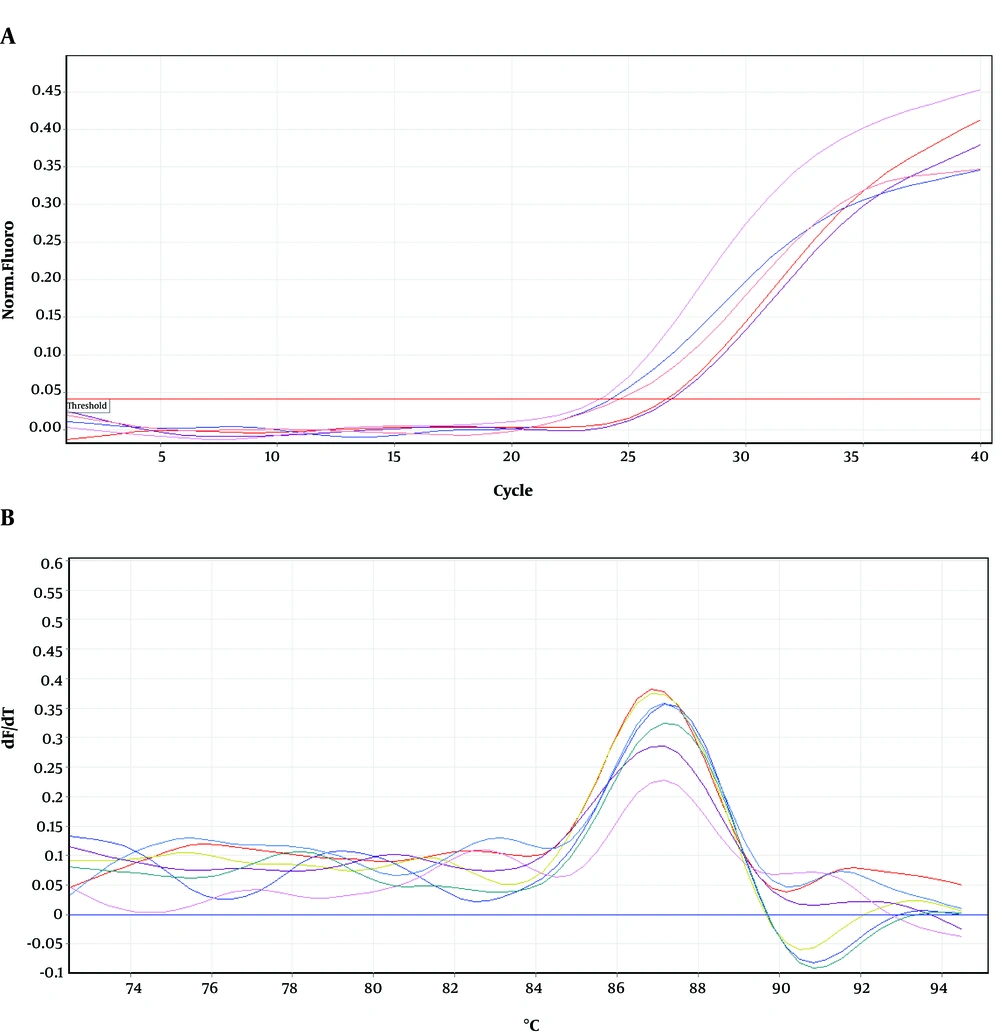
To determine the location of possible mutation in rifampin resistant clones (MN1-MN5), the rifampin resistance-determining region of rpoB gene (containing cluster I and II) was amplified by PCR method and the result of gel electrophoresis can be seen in Figure 1. Then, PCR products were sequenced by DNA sequencing. Although MN2-MN4 were resistant to rifampin, the results of DNA sequencing and comparison with sequence of rpoB gene of MG1655 (ID: 948488) did not showed any change in clusters I and II of RpoB subunit of RNA polymerase (data not shown). However, MN5 had alternation in this region (Figure 2). Ile572 (ATC) was changed to Leu (CTC). Ile572 is a conserved amino acid in rifampin resistance-determining region of rpoB gene among Gram positive and negative bacteria (11). Ile and Leu are hydrophobic amino acids and have similar characteristics. Other clusters, including N and III might have mutation in these clones.
4.4. Ciprofloxacin-induced Mutagenesis in umuC Null Mutant
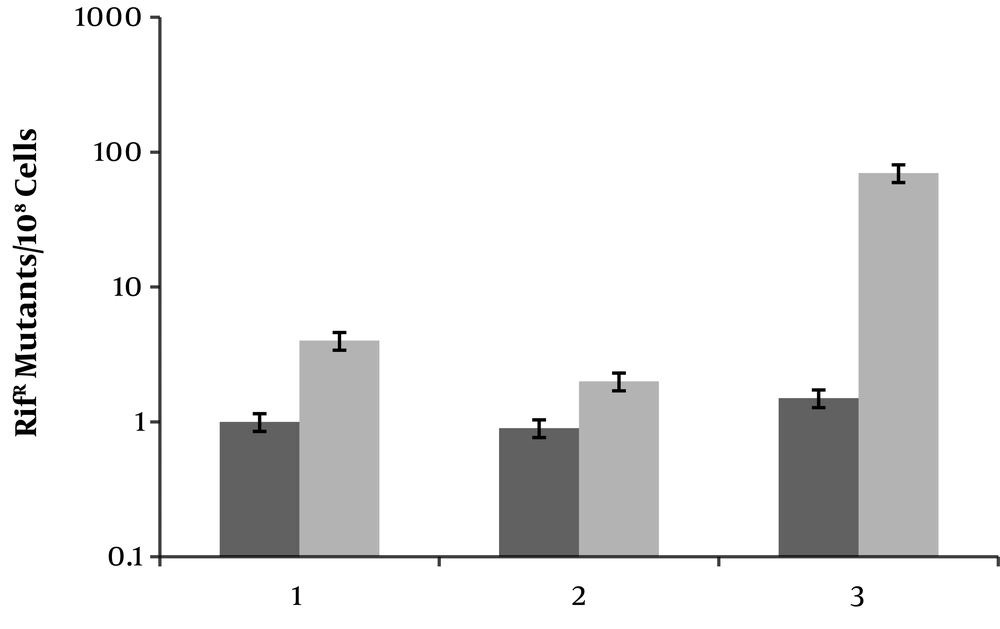
To see whether ciprofloxacin induces mutagenesis in E. coli umuC mutant, strains MG1655 (wild type), JW11731 (umuC) and M2 (ciprofloxacin resistant clone with overactivated AcrAB-TolC efflux pump) were used. Addition of ciprofloxacin (0.1 × MIC) resulted in less than 10 and approximately 100-fold increases in frequency of rifampin resistant mutants in MG1655 and M2, respectively (Figure 3). Interestingly, MN5 produced rifampin resistant clones, but relatively less than MG1655. Thus, ciprofloxacin induces mutagenesis even in the absence of active UmuC.
Frequencies of rifampin resistant (RifR) mutants emerging from different E. coli strains. The ratio of RifR cells were determined in cultures that either untreated (black bar) or treated with ciprofloxacin (grey bar). Lanes 1, 2 and 3 are wild type, MG1655, umuC mutant and M2. Values are the means of three independent experiments ± SD.
4.5. Expression of SOS Genes
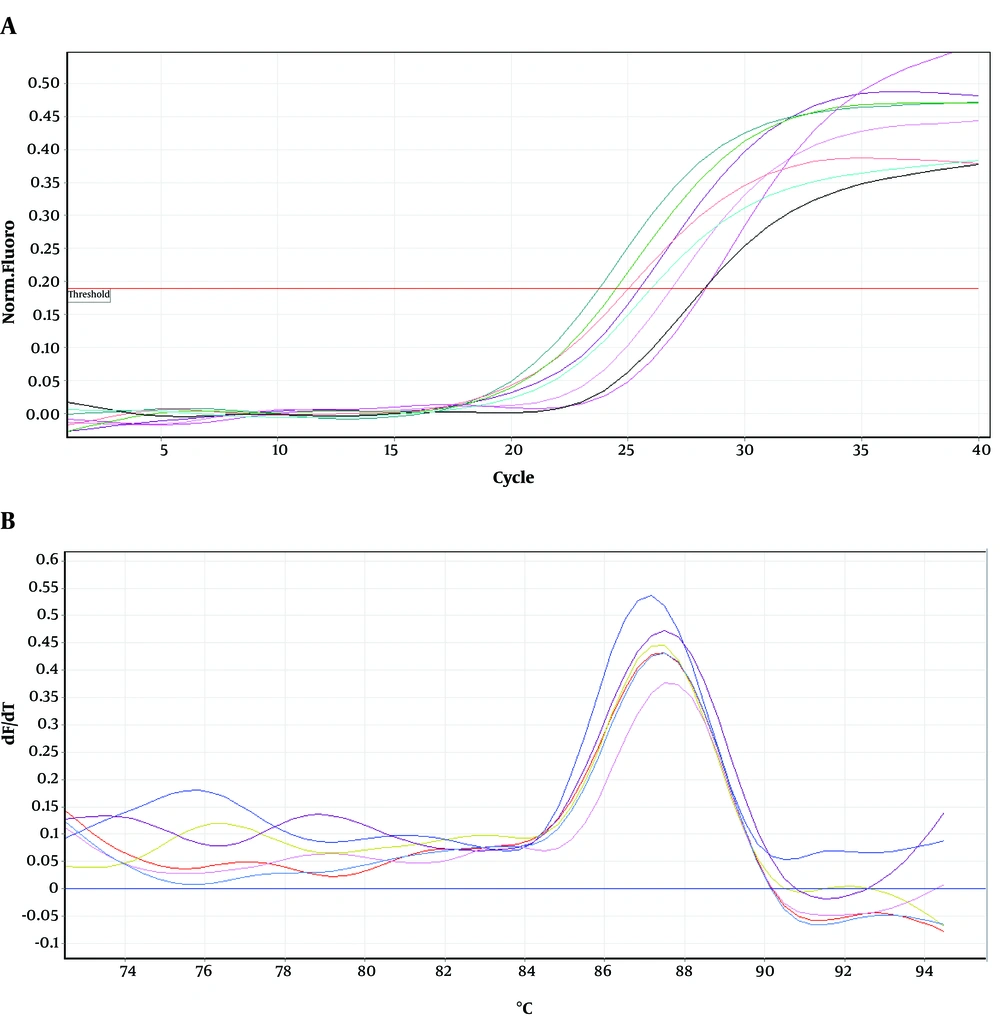
Figures 4-6 show amplification and melting curves of recA, umuD, and dinB genes, respectively. Table 2 shows the relative expression of these genes in different mutants. recA gene was not overexpressed in MN1 with low resistance to ciprofloxacin and rifampin (Table 2). However, this gene was overexpressed in MN2 and MN5 (P < 0.05) with high level of resistance to rifampin (Table 2). Moreover, the expression of umuD and dinB was increased in MN2 and MN5 (P < 0.05), but not in MN1 (Table 2). umuD gene is located upstream of umuC in an operon. Therefore, overexpression of umuD alone did not lead to production of UmuD’2C.
| Strain/Mutant | Expression of Genes | |||||
|---|---|---|---|---|---|---|
| recA | P-Value | umuD | P-Value | dinB | P-Value | |
| MG1655 | 1 | - | 1 | - | 1 | - |
| M2 | 7.1 | 0.02 | 3.54 | 0.03 | 9.4 | 0.01 |
| MN1 | 1.2 | 0. 5 | 1 | - | 1 | - |
| MN2 | 2.5 | 0.05 | 2 | 0.05 | 2 | 0.05 |
| MN5 | 5.4 | 0.02 | 3.8 | 0.02 | 2.5 | 0.05 |
aValues represent fold change (mean of two sets with two samples) in comparison with wild type strain (MG1655). In all cases the standard deviation was less than 10% of mean. Values more than 2 were considered overexpression.
5. Discussion
In E. coli, the mutagenic response to ciprofloxacin is dependent on TLS pathways through SOS-induced DNA polymerases, including pol IV and pol V (UmuD’2C). Inactivation of umuCD inhibits the mutagenic response to ciprofloxacin and prevents the emergence of ciprofloxacin-resistant mutants (3, 23). Despite its important role in mutagenesis and its regulatory effect on the UmuC and pol IV activities, the active umuD gene is not found in the genome of Gram-positive bacteria, including S. pneumonia and S. aureus (5, 14). However, these Gram-positive bacteria show increased mutation frequency in the presence of sub-inhibitory concentrations of ciprofloxacin (16, 22, 24). The mechanism of mutagenesis in these bacteria relies on acquiring mutations in rpoB gene. In this study, we aimed to identify possible alternative mechanisms of mutagenesis to ciprofloxacin in the absence of UmuC in E. coli. We detected rifampin-resistant umuC null mutants, although they were not as common as those in Gram-positive bacteria in the absence of UmuC-like proteins (5, 16, 22). Therefore, the emergence of rifampin-resistant mutations in umuC mutants did not lead to increased mutagenesis following exposure to ciprofloxacin. Also, the low level of mutagenesis in umuC null mutants hinders the formation of clones with high levels of resistance to ciprofloxacin, as we could not produce clones with MICs above 2 µg/mL for ciprofloxacin in this genetic background. However, prolonged contact with ciprofloxacin increased resistance to rifampin, as MN5 showed more resistance to rifampin than MN2-MN4, and additional mutation was found in RpoB.
On the other hand, resistance to ciprofloxacin emerges rapidly in Staphylococcus species, considering its marginal susceptibility to ciprofloxacin. A single mutation in grlA gene (encoding DNA topoisomerase IV subunit A) is sufficient to confer resistance to ciprofloxacin. However, resistance to ciprofloxacin in E. coli, with a significantly higher intrinsic susceptibility to ciprofloxacin, requires multiple mutations, involving the drug target (gyrA and gyrB) and drug accumulation (overactivation of the AcrAB-TolC efflux pump) (9, 25). Activation of the AcrAB-TolC efflux pump occurs after the induction of mutagenic SOS DNA pol V and pol IV (18). Therefore, high levels of resistance to rifampin are expected following the overactivation of the efflux pump, as it was found for the M2 mutant. However, few rifampin-resistant mutants were formed in the umuC background in the absence of DNA pol V. It seems that the presence of other genetic factors and the site of mutation in the β subunit of RNAP are important. This genetic factor may not be MutSL, as rifampin resistant mutants were generated in mutS and mutL genetic backgrounds after exposure to ciprofloxacin (22).
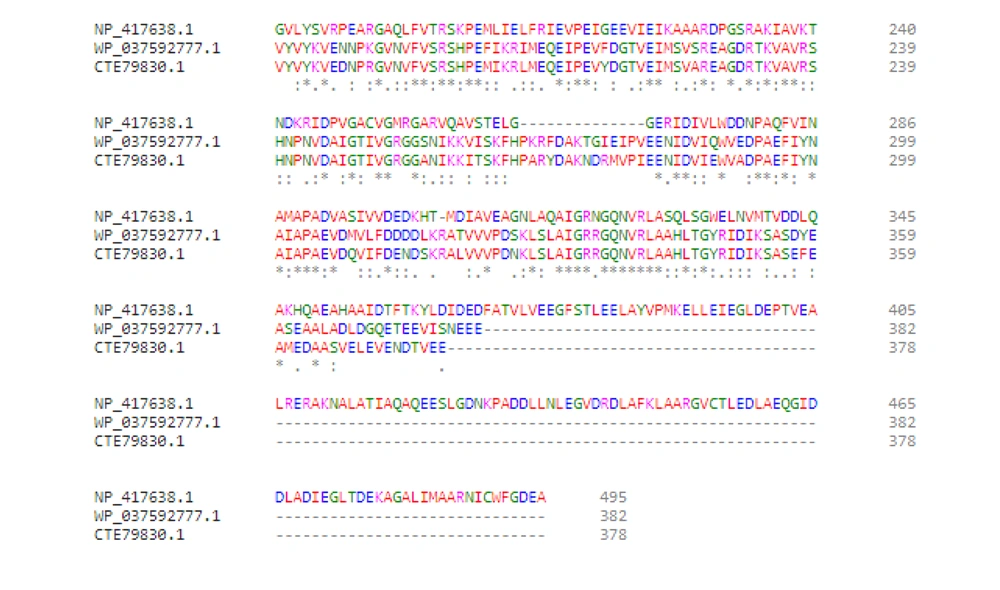
Rifampin interferes with the elongation step of transcription. Besides transcription, progression of DNA replication is inhibited by damage caused by ciprofloxacin. This blockage eventually causes double-strand breaks (DSBs) through formation of either R-loops or single-stranded DNA (4). Generally, DSBs are recognized as potent inducers of SOS response. Adequate amounts of DSBs or R-loops may be found in the umuC clone (MN5), leading to the overexpression of recA, umuD, and dinB. Therefore, formation of RNA polymerase mutants could facilitate the termination of transcription so that the RNA polymerase would not act as a roadblock for DNA replication. Moreover, the overexpression of umuD and dinB genes did not enhance mutagenesis in umuC mutants, as the active UmuD2C was not present, and the possible overproduction of UmuD2 or UmuD’2 prevented the access of pol IV to replisome (6). Therefore, DNA pol IV could not act as the main pathway of mutagenesis in E. coli umuC mutants. However, there is no umuD in Streptococci species, such as S. uberis and S. pneumonia, while dinB is present (almost 42% identical to that of E. coli). Arg38 and Cys66, as conserved residues in DinB, are involved in interactions with RecA (Figure 7) (7). It is possible that DinB acts as the main mutagenic DNA polymerase in these Gram-positive bacteria.
In the present study, we found that most ciprofloxacin-induced mutations in RpoB were not located in cluster I and II, except one at Ile572 (cluster II) in MN5. However, the higher level of resistance to rifampin in comparison with other mutants (MN1-MN4) may indicate that these mutants had acquired a primary mutation either in other clusters of RpoB (clusters N and III) or in other subunits of RNAP (RpoA and RpoC). Also, Ile572 was converted to Leu in MN5. Pourahmad Jaktaji and Nourbakhsh recorded this mutant protein in the National Center for Biotechnology Information (NCBI) database (protein ID: AWH12965.1). Ile and Leu are hydrophobic amino acids with similar characteristics. Replacement of Ile572 with Thr and Asn, as polar amino acids, has been reported in the literature (13). The Ile572Thr and Ile572Asn mutations are categorized as mutations that decrease the transcriptional slippage efficiency (yielding one protein product), relative to wild type RNAP, and also influence the slippage directionality by inhibiting insertions more than deletions. Therefore, mutations at position 572 of RNAP do not seem to result in acquiring different mutant phenotypes.
On the other hand, Pro564Leu mutation increased the slippage efficiency (yielding more than one protein product) and also increased the tendency of RNAP toward insertions rather than deletions (13). This finding was confirmed by Varhimo et al. (16), as they found a variety of mono- to tetra-nucleotide insertions at positions 512, 515, 516, 531, 532, and 564 of RpoB, which might cause diverse mutant phenotypes due to the production of different RNAP mutant enzymes. Therefore, it seems that mutations in clusters I and II of β subunit of RNAP affect (either increase or decrease) the mutation rate in the elongation step of transcription and may affect the interaction of RNAP with proteins, involved in transcription elongation and termination. The substitution of Ile572 with Phe was also detected in RpoB, which might be associated with the suppression of termination defect in nusA mutants (11). It was also found that other mutations, including Ser522Phe, ΔAla523 (deletion of Ala523), Leu524Pro, and Thr563Pro, are suppressors of nusA mutant phenotype (11, 26). It was also found that position 522 in cluster I of RNAP β subunit is prone to acquiring insertion mutations (mono- to tetra-nucleotide insertions) in S. uberis (16), and the slippage phenotypes increased in E. coli (13). Therefore, it seems that there is a relationship between the transcription slippage efficiency of RNAP (mutation rate) and NusA activity.
Moreover, it was found that the single nusA11 (dinB+) mutant contains wild type E. coli level of mutagenesis to UV light, indicating that SOS induction and DNA pol V are functional (27). Also, double nusA-dinB mutants exhibit the same level of mutagenesis, suggesting the essential presence of intact NusA, since in the absence of DinB, NusA can interact with DNA pol V. However, it is not clear whether a mutation in nusA or dinB would decrease the mutation rate in S. uberisumuC mutants. It was proposed that NusA interacts physically with DinB (28). The C terminal 263 amino acids of NusA (from amino acid 233 to amino acid 495), which bind to RNAP, seem to be important for interaction with DinB; however, the residues of DinB for the interaction with NusA are not clear (8). There are differences in NusA between E. coli and Streptococci species (S. uberis and S. pneumonia). Generally, NusA of E. coli is longer than that of Streptococci species (Figure 8). However, there are conserved residues among them, which may interact with DinB; we did not evaluate the sequence of nusA gene for possible mutations. Wild type NusA recruits DinB to sites where RNAP is stalled by a gap in the transcribed strand. If DinB cannot be recruited due to the presence of UmuD2 or UmuD’2, the presence of intact or mutant nusA may not be important. However, the question arises as to how mutations in cluster I and II RpoB can help bacterial cells rescue stalled RNAP.
5.1. Conclusions
In conclusion, to increase the efficiency of ciprofloxacin against Gram-negative bacteria, it is necessary to inactivate umuC gene. Inactivation of umuC did not inhibit the generation of rifampin resistant mutants. This did not increase mutagenesis. Thus, finding an inhibitor of umuC, but not an inhibitor of the umuCD operon is suggested. It can be used as an adjuvant along with ciprofloxacin to eliminate infection caused by E. coli.