1. Background
Human immunodeficiency virus (HIV) infection causes lifelong viral persistence in infected individuals (1). Human immunodeficiency virus,, human T-cell leukemia virus type 1 (HTLV-1), hepatitis B virus (HBV), and hepatitis C virus (HCV) are four sexually transmissible blood-borne viruses that cause life-threatening diseases. In HIV-infected cases, co-infection with these tumor viruses may worsen the clinical progression of associated diseases such as hepatocellular carcinoma (HCC) and adult T-cell leukemia/lymphoma (ATL) (2, 3).
Human immunodeficiency virus co-infection with HTLV-1, as an oncogenic retrovirus, should be considered more seriously in the endemic areas because of controversial results reported ranging from protective to aggressive impacts in different endemic regions (4). The northeast of Iran, mainly Razavi Khorasan province, is an endemic region for these four viruses (2, 5). At present, according to the United Nations Program on HIV/AIDS (UNAIDS) update in 2020, 59,000 [33,000 - 130,000] cases of HIV positive cases are living in Iran with an incidence of around 0.05 [0.01 - 0.15] per 1,000 population (6). Some studies in central Iran demonstrated that circulating HIV-1 belonged to the CRF35-AD and CRF01-AE recombinant forms of the M group (7). However, in the northeast of Iran, Naderi et al. reported that the circulating subtype is Uganda and Kenya subtype A (8). HIV infection causes lifelong viral persistence in infected individuals (1). Therefore, reliable methods for the effective monitoring of clinical status and antiviral therapy should be considered more seriously.
The best markers for monitoring the HIV-infected subjects are the evaluation of the CD4+ T-cell, HIV reservoir, and HIV viral load to trace the behavior of the virus activity and host (9). However, in the poor-stricken area, the total lymphocyte count and hemoglobin concentration can act as surrogate markers that can be used for monitoring HIV-infected subjects (10). The CD4+ T cells participate in numerous operations to gain immunity against viruses (11). Moreover, CD8+ T cells have key roles in the immune response of the cells and control of HIV replication in the early phase of infection. The CD4+ T cell count is a standard method for the immunological evaluation of HIV-infected cases, which can be fulfilled utilizing flow cytometry (12, 13).
Since CD4+ T cell count may fluctuate with intercurrent illness and physiological and test variability, a single measurement is not efficient for clinical management. Moreover, the CD4+ T cell numbers in patients are lower in some regions of Asia and Africa than that in Europe (14). The quantification of HIV RNA in plasma has been introduced for determining the disease progression and the beginning of treatment time (15, 16). Besides, a diversity of sophisticated molecular techniques, such as TaqMan Real-Time PCR (qRT-PCR) and COBAS Amplicor monitor test, are applied for the quantitation of the viral load (17-19). The clinical variability and phenotypic heterogeneity in clinical manifestations of HIV-related symptoms and the duration of latency in a given population are surprisingly high (20). The HIV-positive subjects travel the whole spectrum from carrying the healthy latent infection to opportunistic infections, and finally severe disease. Therefore, it is necessary to make a very precise prognosis method to obtain the best estimation for starting the therapy and monitoring the treatment.
Recent studies have demonstrated that the outcome of the HIV infection probably depends on the virus and host interactions in the context of other viral co-infections such as herpes simplex virus (HSV), HTLV-1, and Kaposi's sarcoma-associated herpesvirus (KSHV) (20). Therefore, with a better understanding of the connections between infectious viral diseases and the epigenetic events, opportunities arise for therapeutic solutions, particularly as epigenetic processes can be reversed.
2. Objectives
In this study, the co-infection of HIV with the HBV, HCV, and HTLV-1, the simultaneous assessment of the HIV- infected host factors, WBC, CD4+, and CD8+ count, along with main HIV factor, viral load using flow cytometry, TaqMan Real-Time PCR, and COBAS-Amplicor monitor test were evaluated for estimation of the progression of infection and also its correlation with the clinical status.
3. Methods
The study included 20 patients with HIV, who were referred to the Triangular Clinic at Mashhad University of Medical Sciences (MUMS), Mashhad, Iran. This clinic is the only therapy center for HIV-positive patients in the Khorasan provinces. A demographic and clinical questionnaire was filled during the study. The characteristic data for the subjects are summarized in Table 1. The first evaluation was carried out at the time of HIV diagnosis, and the last one was performed in a one-year interval. The clinical staging was assigned by an infectious disease specialist based on the revised criteria of the World Health Organization (WHO) staging system at both time-points (21).
| Gender | Age | Transmission Route | Clinical Stage | Duration of Infection (Year/Month) |
|---|---|---|---|---|
| Female | 40 | Needle Stick | Stage 4 | 5/08 |
| Male | 45 | Tattoo | Stage 3 | 9/6 |
| Male | 38 | Tattoo | Stage 4 | 4/08 |
| Male | 49 | IV drug using | Stage 3 | 2/6 |
| Male | 45 | IV drug using | Stage 3 | 11/3 |
| Male | 30 | IV drug using | Stage 2 | 2/8 |
| Male | 28 | IV drug using | Stage 1 | 0/4 |
| Male | 50 | IV drug using | Stage 3 | 11/6 |
| Male | 38 | IV drug using | Stage 4 | 1/6 |
| Male | 33 | IV drug using | Stage 1 | 0/8 |
| Male | 33 | IV drug using | Stage 2 | 0/6 |
| Male | 48 | IV drug using | Stage 3 | 11/0 |
| Male | 22 | IV drug using | Stage 1 | 0/1 |
| Male | 36 | IV drug using | Stage 2 | 1/9 |
| Male | 29 | IV drug using | Stage 1 | 2/4 |
| Male | 47 | IV drug using | Stage 4 | 6/2 |
| Female | 25 | Sexual contact | Stage 2 | 3/6 |
| Male | 34 | Sexual contact | Stage 1 | 6/8 |
| Female | 25 | Sexual contact | Stage 3 | 1/8 |
| Female | 33 | Sexual contact | Stage 3 | 8/4 |
The clinical evaluations, interviews, serology tests, and CD4+ cell counts were repeated every six months. Routine laboratory tests, including the complete blood count and differential cell counts were assessed. Moreover, serologic tests for the detection of HCV, HTLV-1, and HBV co-infections were carried out. We assessed HCV and HTLV-1 infections by a commercial third-generation ELISA kit (Diapro, Italy), HBsAg using an available ELISA kit (Radim, Italy), and HIV infection by a one-step ELISA kit (HIV Uniform II Ag/Ab, Biomerieux, The Netherlands) according to the manufacturers’ instructions. Human immunodeficiency virus infection was confirmed by an immunoblot commercial kit (INNO-LIA HIV I/II Score, Innogenetics, Belgium). The seropositive results for HTLV-1 and HCV were confirmed using the PCR method as previously described (2).
Peripheral blood mononuclear cell isolation was collected from the participants. The peripheral blood mononuclear cells were extracted by Blood mini kit (Qiagen, Germany). Plasmas were frozen immediately after separation and stored at -70°C until analyzed. The flow cytometry assay was carried out by direct staining of cells conjugated or isotype control antibodies. Three color antibodies (CD4-FITC, CD8 R-PE, and CD3-CYQ) were used for cell staining (IQ Products, The Netherlands). The samples were analyzed using a FACSCalibur (BDBiosciences, USA) flow cytometer.
3.1. Human Immunodeficiency Virus RNA Assays
Total RNA was extracted from 200 to 400 µL of plasma using the QIAamp blood kit (QIAGEN, Inc., Valencia, CA). The viral load was assessed via two different quantitative techniques—real-time TaqMan using a commercial TaqMan kit (Qiagene, Germany) by a Q 6000 machine (Qiagen, Germany) in the Ghaem Hospital Virus Laboratory (Mashhad, Iran) and quantitative COBAS Amplicor HIV monitor test V,1.5 (Roche, Germany) in Kivan Virus Medical Laboratory (Tehran, Iran). For phylogenetic analysis, extraction of viral RNA from serum was performed using High Pure Nucleic Acid Purification kit (Roche-Germany) according to the kit protocol. One-step RT-PCR reaction was performed using one-step RT PCR Superscript III (Invitrogen). The partial of env gene from the genomic region of c2-v5 (6937-7651) region according to reference numbering virus HXB-2 amplified by nested PCR with the following primers: 5'ATG GGA TCA AAG CCT AAA GCC ATG TG3' and 5'AGT GCT TCC TGC TGC TCC CAA GAA CCC AAG3' to the first step and 5' CCT CAG CCA TTA CAC AGG CCT GTC CAA AG3'and 5'TTA CAG TAG AAA AAT TCC CCT C3' in a second step.
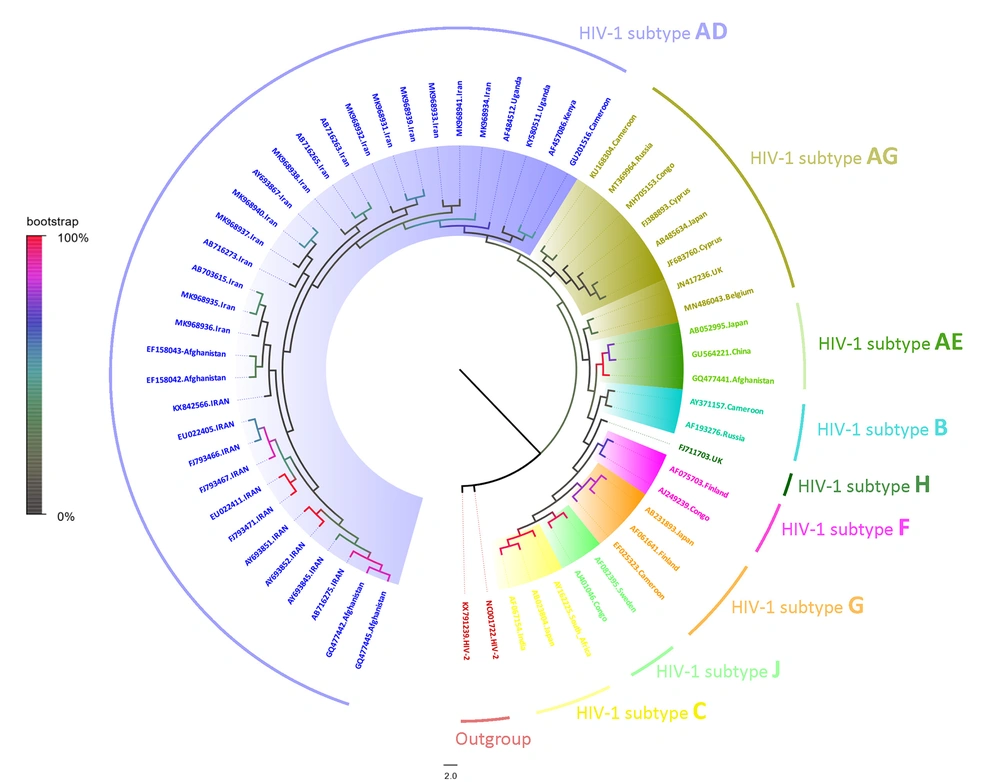
The amplified fragments were carried out by direct sequencing assay according to a protocol described in the ABI Prism Dye Terminator Cycle Sequencing Ready Kit (PerkinElmer Cetus, Norwalk, CA, USA). The nucleotide sequences were aligned by Clastal W using the BioEdite software (v 7.0.9.1). The reference sequences were obtained from the Gene bank (www.ncbi.nlm.nih.gov) and HIV database (www.hiv.lanl.gov). The phylogenetic tree was constructed by MEGA software (version X) with the maximum-likelihood method and GTR substitution model with 1000 bootstrap values (Figure 1) (GeneBank accession: MK968931-41) (22). The data were analyzed using SPSS software (version 13.0; SPSS Inc, Chicago). The outcomes were considered statistically significant if the P-value was < 0.05. Spearman’s rank correlation coefficient was used to compute the correlations between variables. The qRT-PCR and COBAS-Amplicor were compared using the Mann-Whitney test.
Phylogenetic analysis of the partial sequences corresponding to the region of C2-V5 in HIV-1 env gene, based on the maximum likelihood method with 1000 bootstrap replicates. The samples analyzed in the present study are shown with MK968931-41 GenBank accession numbers, and all other representative reference sequences from nine HIV-1 subtypes were obtained from GenBank (https://www.ncbi.nlm.nih.gov/genbank).
4. Results
4.1. Demographic and Clinical Characterization of the Study Population
The mean age of subjects (16 males and 4 females) was 36.4 ± 8.6 years (age range: 22 - 50 years). Furthermore, 13 individuals were intra-venous drug users (IVDUs), four had sexual contact with an HIV-seropositive case, two had a tattoo, and only one was infected through a needle stick (occupational infection). Table 1 shows the demographic, clinical, and laboratory information of the subjects studied.
4.2. Human Immunodeficiency Virus Co-infections
The majority of patients were IVDUs. Therefore, the co-infections with HBV and HCV were too high. The HIV-HCV co-infection was seen in 75% (15/20), and the co-infection with HBV was found in 75% of cases. The HTLV-1 co-infection with HIV was detected in 15% (3/20) of subjects who also had HBV and HCV infections. Table 2 shows the results of the co-infections.
| WBC Count | First CD4 Count | Last CD4 Count | First CD8 Count | Last CD8 Count | Viral Load | Co-infections | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cobas Amplicor | Real Time PCR | HTLV-1 | HBV | HCV | ||||||||
| Copies/ml | Log | Copies/ml | Log | |||||||||
| 1 | 3700 | 281 | 412 | 810 | 341 | 100 | 2 | 100 | 2 | Negative | Positive | Positive |
| 2 | 3600 | 258 | 150 | 650 | 311 | 165000 | 5.22 | 156105 | 4.97 | Negative | Positive | Positive |
| 3 | 4300 | 69 | 381 | 603 | 495 | 100 | 2 | 100 | 2 | Negative | Positive | Positive |
| 4 | 3900 | 465 | 400 | 285 | 383 | 819 | 2.91 | 9789 | 3.77 | Negative | Negative | Negative |
| 5 | 4400 | 459 | 290 | 407 | 703 | 3850 | 3.59 | 1065 | 2.81 | Negative | Positive | Positive |
| 6 | 3500 | 292 | 130 | 1479 | 709 | 480000 | 5.68 | 291795 | 5.24 | Positive | Positive | Positive |
| 7 | 7400 | 485 | 651 | 1275 | 1432 | 45400 | 4.66 | 21032 | 4.10 | Positive | Positive | Positive |
| 8 | 6500 | 660 | 534 | 1663 | 800 | 13400 | 4.13 | 13014 | 3.89 | Negative | Positive | Positive |
| 9 | 3700 | 157 | 133 | 767 | 462 | 43800 | 4.64 | 6711 | 3.60 | Negative | Positive | Positive |
| 10 | 3900 | 407 | 407 | 930 | 930 | 43100 | 4.63 | 57996 | 4.54 | Negative | Positive | Positive |
| 11 | 6600 | 752 | 558 | 401 | 812 | 2300 | 3.36 | 5595 | 3.53 | Negative | Positive | Positive |
| 12 | 5700 | 762 | 593 | 932 | 866 | 4220 | 3.63 | 3248 | 3.29 | Negative | Positive | Positive |
| 13 | 8900 | 243 | 251 | 860 | 720 | 134000 | 5.13 | 680000 | 5.61 | Negative | Positive | Positive |
| 14 | 3900 | 20 | 25 | 895 | 675 | 43600 | 4.64 | 122918 | 4.87 | Negative | Negative | Negative |
| 15 | 4400 | 359 | 561 | 473 | 841 | 100 | 2 | 100 | 2 | Negative | Negative | Negative |
| 16 | 4000 | 659 | 491 | 452 | 357 | 2790 | 3.45 | 658 | 2.60 | Negative | Negative | Negative |
| 17 | 8900 | 653 | 653 | 625 | 625 | 41700 | 4.62 | 24139 | 4.16 | Negative | Positive | Positive |
| 18 | 4000 | 118 | 112 | 700 | 898 | 173000 | 5.24 | 201000 | 5.08 | Negative | Negative | Negative |
| 19 | 6100 | 601 | 475 | 1068 | 590 | 26800 | 4.43 | 164014 | 4.99 | Positive | Positive | Positive |
| 20 | 5000 | 465 | 413 | 1284 | 936 | 334000 | 5.52 | 602527 | 5.56 | Negative | Positive | Positive |
4.3. Viral Load Quantification
Human immunodeficiency virus viral load quantification results were expressed as copy number of HIV/mL and log10 of the copy number. The average copy number of HIV by qRT-PCR and Cobas-Amplicor monitor test was 70902 ± 26527 copies/ml and 77948.95 ± 28375 copies/ml, respectively. As Table 3 shows, there was a strong correlation between the two methods for assessing HIV copy number (R = 0.881, P < 0.0001). As no significant difference was observed between these methods, it could be concluded that they have the same reliability.
| Factor | Cobas Amplicor HIV Monitor Test(Copy Number) | qRT-PCR (Copy Number) | ||
|---|---|---|---|---|
| Correlation | P-Value | Correlation | P-Value | |
| CD4+ | -0.416 | 0.068 | -0.372 | 0.106 |
| CD8+ | 0.327 | 0.160 | 0.292 | 0.211 |
| CD4+/CD8+ ratio | -0.691 | 0.001 | -0.539 | 0.014 |
| Cobas amplicor HIV monitor test (copy number) | - | - | 0.881 | <0.0001 |
| WBC count | -0.089 | 0.709 | 0.068 | 0.776 |
| Duration of infection | -0.295 | 0.206 | -0.318 | 0.172 |
| HTLV-1co infection | 0.547 | 0.013 | 0.401 | 0.079 |
4.4. Phenotyping
Results of the flow cytometry analyses showed that the first and last absolute counts of CD4+ T cells were 428.68 ± 213.18 cells/mm3 and 399.73 ± 178.21 cells/mm3, respectively. These measures for CD8+ T cells were 375.49 ± 827.95 cells/mm3 and 694.30 ± 268.40 cells/mm3, respectively. The ratio of CD4+ to CD8+ T cells at the time of diagnosis was 0.629 ± 0.53 and in the last evaluation was 0.608 ± 0.35. There were no significant differences in CD4+ absolute counts and CD4+ to CD8+ ratio between the first measurement and the last one. A meaningful inverse correlation was detected between the first CD4+ count and the clinical stage (R = -0.60, P = 0.006). Moreover, there were significant inverse correlations among the first CD4+ and first CD8+ count with duration of the infection (R = -0.98, P = 0.03 and R = -0.52, P = 0.018, respectively).
4.5. Correlations of Viral and Host Indices
A significant inverse correlation was observed between the last CD4+ T cell count and the copy number obtained by Cobas-Amplicor HIV monitor test results (R = -0.41, P = 0.06). Furthermore, inverse correlation coefficients were found between CD4/+CD8+ ratio and the copy number obtained by Cobas-Amplicor HIV monitor test (R = -0.691, P = 0.04), and between CD4+/ CD8+ ratio and qRT-PCR (R = -0.539, P = 0.01). Conversely, a direct correlation was observed between CD8+ value and HTLV-1 co-infection (Table 3).
4.6. Phylogenetic Analysis
Phylogenetic analysis of and c2-v5 in env genes demonstrated that all the HIV-infected subjects are located within clad of AD (Gene Bank accession: MK968931-41). The results of phylogenetic trees confirmed each other.
5. Discussion
Measuring the HIV viral load in the plasma can be useful for the determination of disease progression and treatment strategy (23). The analogy between the results obtained from different assays is momentous, which can help to make the best decision for choosing the assay of analysis. CD4+ T-cells, the main targets of HIV infection, have a central role in orchestrating cellular immune reactions. These cells activate anti-HIV specific CD8+ T cells involved in viremia control (24, 25). Measurement of the CD4+ T cell count is a standard method for the evaluation of HIV-infected patients because of the close correlation with the clinical manifestations of HIV infection. In our study, 75% of the subjects had co-infection with HBV and HCV; this high amount of co-infection seems to be associated with the sharing of syringes in drug abusers, as most of our patients were IVDUs. Hepatitis B virus and HCV infections are extremely common among HIV-infected patients because of their common transmission routes (26, 27). The HCV infection causes an increase in the HIV/AIDS epidemic due to the facilitation of HIV progression (28). However, the treatment of HIV with antiretroviral therapy (ART) in the HIV/HCV co-infected patients leads to a delay in the cirrhosis progression (29).
The co-infection of HIV with HTLV-1 was lower (15%). However, these subjects were also drug abusers and had HBV and HCV infection too. It seems that the transmission rate of HTLV-1 is less than the HBV and HCV, as we have previously reported in the drug abusing prisoners in our endemic region (2). Although HITLV-1 has the same prevalence as HBV and HCV in this region (2) and despite the same route of transmission, the rate of HBV/HCV infection is higher (5 times) than HTLV-1 in HIV-infected patients. It is well-known that HTLV-1 is mainly transmitted from mother to neonate via breastfeeding. However, in any route of infection in vivo, HTLV-1 mainly transmitted through cell-cell contacts via a very specialized cell compartment called "virological synapse". Therefore, in contrast to HCV, HBV, and HIV, since there is no HTLV1 free version in body fluids, they are not infective. Human T-cell leukemia virus type-I and HIV as human retroviruses infect CD4+ T cells. There has been a direct relationship between HIV viral load and the clinical progression. Thus, high HIV viral load in HTLV-1 co-infection has been implicated in clinical progression and severity of AIDS (4). Rahimi et al. in the northeast of Iran have shown that HIV viral load in HTLV-I/HIV co-infected subjects was higher than HIV-infected patients, and HTLV-I proviral load in HTLV-I/HIV co-infected subjects was lower than in HTLV-I infected patients. Therefore, the associations between HIV-HTLV-1 co-infection and clinical progression of HIV/HTLV-1 related diseases emphasize that in the endemic infected regions, more attention should be paid to these patients.
The present study showed a direct correlation between the results of the two common tests, including qRT-PCR and Cobas-Amplicor monitor test. Therefore, it was concluded that these techniques confirm each other. The phenotypic heterogeneity in clinical manifestations of the HIV-related symptoms and the duration of latency in a given population are surprisingly high. Hence, HIV-positive subjects travel the whole spectrum from carrying the healthy latent infection to opportunistic infections, and finally severe disease. The study revealed a considerable negative correlation between CD4+ count and the copy number obtained from the Cobas-Amplicor HIV monitor test. This outcome was expected as the CD4+ T cells are the main target of HIV. Therefore, their levels decrease in HIV-infected patients. However, this decrease was not detected by the application of qRT-PCR.
Although no correlation was found between the CD8+ count and the copy number, remarkable correlations were found between the CD4+/ CD8+ ratio and the two techniques. Therefore, this parameter can be considered more significantly to trace HIV infection. Since three-quarters of the HIV-infected patients were also infected by HBV and HCV due to sharing drug needles, this group of patients should be given more attention. The practical proceedings could include providing the syringe for drug users, increasing the education level, and facilitating access to health services. Moreover, the patients infected by one or two of the above-mentioned viruses should care more carefully because they probably have a high risk of infection from other viruses. The results of this study demonstrated that the HIV infection follow-up could be carried out by either the TaqMan RT-PCR or the Cobas-Amplicor HIV monitor test. However, the Cobas-Amplicor HIV monitor test is more reliable. Moreover, the CD4+/CD8+ ratio obtained by flow cytometry can confirm the results of the aforementioned two tests. Furthermore, there was a decreasing trend in the CD4+ level among subjects in stages 2 and 3. However, this specific trend was not observed among patients in Stages 1 and 4.
In the present study, the phylogenetic analysis showed that HIV-1 belonged to the AD genotype recombinant form of the M group, that is consistent with other studies from Iran. The first study by Naderi et al. on HIV-1 IVDUs positive subjects in 2005 demonstrated the presence of subtype A in Khorasan province (8). They suggested African origins, Uganda and Kenya, for the HIV circulating in this large region. However, the phylogenetic relationship findings in the present study showed radical changes to the CRF35-AD, which is the predominant subtypes in other parts of Iran (22).
There are several important reasons for investigating genetic subtypes of HIV-1 circulating in an area. First, different subtypes express variant envelope proteins, and this is likely to affect vaccine development. For developing an effective vaccine, it is important to know geographically and/or subtype-specific strains (30). Second, the genetic variation among subtypes can lead to problems in some detection methods (31). Third, several studies have tried to correlate genetic variability to biological properties, such as transmission, disease prognosis, and response to treatment (32-34). For example, it has been suggested that HIV-1 subtype E, now recognized as CRF01-AE, may be more transmissible than subtype B in Thailand (33, 35, 36). Fourth, knowledge about circulating subtypes is essential for understanding the epidemiology and spread of the HIV-1 pandemic (37, 38).
5.1. Conclusions
One of the objectives of this study was to investigate the relationship between different subtypes circulating in Iran with the biological characteristics of the virus. Because only one subtype has been identified in this study, which is consistent with the results of previous studies in Iran, this aim is unattainable and requires a study with a larger number of samples.