1. Background
Antimicrobial resistance (AMR) due to ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa, and Escherichia coli) is posing a serious threat to public health globally (1, 2). According to the WHO, carbapenem-resistant A. baumannii (CRAB) has been one of the main concerns for the last 10 years due to the risk of antibiotic resistance. Acinetobacter baumannii is considered a “red alert” pathogen by the Infectious Diseases Society of America due to its resistance against antibiotics. It is associated with major health concerns in different regions of the world. These infections are responsible for about 1.5 million cases yearly (3, 4).
Acinetobacter baumannii is one of the most prevalent pathogens causing nosocomial infections, especially in people admitted to the intensive care units (ICUs) (5, 6). It has been demonstrated that such resistance in these pathogens is due to plasmids, transposons, and integrons, particularly class I and class II. The nosocomial propagation of isolates has been illustrated in both environmental and clinical specimens (7, 8). The development of resistance in CRAB pathogens has been increasing due to the emergence of class B, C, and D carbapenemase, which declines membrane permeability, modifies PBP, and increases efflux pump expression. Carbapenem resistance in A. baumannii is mainly mediated by intrinsic (OXA-51) or acquired (OXA-23) oxacillinases (9, 10). It is considered a health care pathogen mostly encountered in several serious medical problems such as septicemia, meningitis, bacteremia, ventilator-associated pneumonia, endocarditis, and urinary tract infection (4, 11). Epidemiologically reported data provide evidence of A. baumannii infectious globally, e.g., in Korea, Iran, Brazil, America, Europe, China, Iraq, Hong Kong, Taiwan, and Argentina. In several regions of the world, due to climate change, community-acquired pneumonia is due to this infection, as reported in the literature (12).
Resistance in ESKAPE pathogens E. faecium (13), S. aureus (14), K. pneumonia (15), A. baumannii (16), P. aeruginosa (17), and E. coli (18) is caused by the enzymatic degradation of antibiotics, target site mutations/modifications, decreased porin expression, and overexpression of multidrug efflux pumps. However, lactamases, such as carbapenem hydrolyzing class D-lactamases (CHDLs) and Metallo-lactamases, are frequently involved in carbapenem resistance. Resistance to CHDLs, also known as oxacillinases, is primarily achieved by the generation of carbapenemase enzymes encoded by the genes of blaOXA-23, blaOXA-40, and blaOXA-58 lineages; however, blaOXA-23 is the most widespread one worldwide (19). In A. baumannii, transposable elements such as insertion sequences (ISAba1) play a key role in carbapenem resistance, since they are found upstream in the promoter regions of the blaOXA-23, blaOXA-40, blaOXA-58, and blaOXA-51 genes, inducing the overexpression of these resistance genes (20, 21).
2. Objectives
The main objectives of this cross-sectional study were to find out the AMR patterns in A. baumannii isolates and evaluate the AMR genes (blaOXA-23, blaOXA-24, blaOXA-40, blaOXA-51, blaOXA-58, blaNDM1, blaIMP, blaVIM, ISAba1, ssul1, sul2, armA, and PER-1) in these isolates from patients admitted to the surgical ICUs at different hospitals in Lahore, Pakistan.
3. Methods
3.1. Sample Collection, Isolation, and Identification of Acinetobacter baumannii
A total of 593 clinical specimens of wound, blood, burn, and pus was collected from different local hospitals from June 2017 to March 2019 from patients admitted to the surgical ICUs in Lahore General Hospital (n-396), Mayo Hospital (n-103), and Jinnah Hospital (n-94), Lahore, Pakistan, using a simple random sampling technique. After collection, the samples were inoculated on blood and MacConkey agar using a disposable wire loop as primary, secondary, and tertiary streaking, and then the Petri plates were incubated at 37°C for 24 h. After the incubation period, the plates were observed for the appearance of bacterial colonies. Acinetobacter baumannii showed non-lactose fermenter colonies on MacConkey agar. The isolated bacterial colonies were then processed for biochemical identification of bacteria using standard biochemical tests (API) 10S kit (Biomeurix) (5).
3.2. Antimicrobial Susceptibility Testing
Antimicrobial Susceptibility testing (AST) was performed by the Kirby-Bauer disc diffusion method according to the Clinical Laboratory Standard Institute (CLSI) guidelines 2020 (3). To standardize the inoculum density for susceptibility tests, McFarland standards were used (10). They were prepared by using different concentrations of barium chloride (BaCl2) and sulfuric acid (H2SO4) to make 0.5, 1, and 2% standards for visual differences. Different concentrations of BaCl2 and H2SO4 were used and stored at 4°C for further use. A fresh colony was picked by a sterile inoculating loop and suspended in 2 mL of normal saline optically equal to 0.5 McFarland standards and streaked by a swab over the entire surface in three to four planes by rotating the plate at 60 °C each time to ensure the even distribution of inoculum. In the end, the rim of agar was swabbed, and the plate was left undisturbed for 15 min to absorb the excess inoculum over it. Finally, antibiotic disks were placed on the inoculated agar plates and incubated at 37°C for 24 h.
Tested isolates were used for AST to a panel of 15 different antibiotics as suggested by CLSI guidelines 2020. The antibiotics were amikacin (30 µg), ceftriaxone (30 µg), ceftazidime (30 µg), cefepime (30 µg), ciprofloxacin (5 µg), colistin (10 µg), co-trimoxazole (23.75 µg), gentamicin (10 µg), imipenem (10 µg), meropenem (10 µg), piperacillin-tazobactam (10 µg), tigecycline (15 µg), polymixin-B (300 units), tobramycin (10 µg), and tetracycline (30 µg).
3.3. Amplification and Detection of Resistance Genes
All of the A. baumannii isolates were processed for DNA extraction using a DNA extraction kit (WizPrepTM). The quality of DNA was checked by agarose gel electrophoresis. Then, 1% agarose gel was prepared in TAE buffer. After mixing and boiling, 0.5 mg/mL of ethidium bromide was added. The mixture was then poured into a casting tray, and a comb was inserted into it. The casting tray was left at room temperature until getting solidified. The seal and the comb were removed carefully, and the gel was placed in an electrophoresis chamber containing TAE buffer. Then, 1 µL of Thermo scientific 6X loading dye was mixed with 6 µL of sample, and a volume of 5 µL was loaded in each of the wells. The first well was loaded with Thermo scientific DNA ladder, and the remaining wells were loaded with the DNA of our interest. In the end, the lid was placed on the gel box, and electrodes were connected with it, and the gel was run at 70 volts for 30 min. The lid of the gel box was removed, and the gel was picked out from the tray using sterile gloves and placed in a gel documentation system (BioRad, Germany).
The resistance genes (blaOXA-23, blaOXA-24, blaOXA-40, blaOXA-51, blaOXA-58, blaNDM1, blaIMP, blaVIM, ISAba1, ssul1, sul2, armA, and PER-1) were amplified using gene-specific primers as given in Table 1 and amplification conditions listed in Table 2. The PCR products were loaded on an agarose gel to visualize the amplified genes using 2% agarose gel. To amplify each gene, a PCR was carried out in a final volume of 25 µL containing 1× PCR buffer, 1U Taq polymerase, 1.5 mM MgCl2, 200 µM of dNTP, 10 pmol of each primer, and 1 µL of extracted DNA. The conditions of amplification were programmed in Master-cycler Eppendorf as follows: Initial denaturation at 94°C for 3 min, 35 cycles of 94°C for 45 s, annealing varying according to the individual gene for 45 s, extension at 72°C for 1 min, and final extension at 72°C for 5 min. The PCR products were separated on the 1.5% agarose gel by electrophoresis, stained with ethidium bromide, and then visualized under a UV gel documentation system (Sigma-Aldrich) (Table 1).
| Gene | Sequence (5’ - 3’) | Amplicon Size, bp | Annealing temperature, °C | Reference |
|---|---|---|---|---|
| blaOXA-51 | F-TAATGCTTTGATCGGCCTTG | 353 | 52 | (22) |
| R-TGGATTGCACTTCATCTTGG | ||||
| blaOXA-23 | F-GATCGGATTGGAGAACCAGA | 501 | 52 | (22) |
| R-ATTTCTGACCGCATTTCCAT | ||||
| blaOXA-24 | F-CAAGAGCTTGCAAGACGGACT | 420 | Not detected | (23) |
| R-TCCAAGATTTTCTAGCTTATA | ||||
| blaOXA-58 | F- AAGTATTGGGGCTTGTGCTG | 599 | Not detected | (24) |
| R- CCCCTCTGCGCTCTACATAC | ||||
| blaOXA-40 | F-GGTTAGTTGGCCCCCTTAAA | 246 | 52 | (25) |
| R-AGTTGAGCGAAAAGGGGATT | ||||
| blaNDM1 | F- GGTTTGGCGATCTGGTTTTC | 621 | 52 | (25) |
| R- CGGAATGGCTCATCACGATC | ||||
| blaIMP | F- GTTTATGTTCATACWTCG | 432 | 48 | (24) |
| R- GGTTTAAYAAAACAACCAC | ||||
| blaVIM | F- TTTGGTCGCATATCGCAACG | 500 | 66 | (25) |
| R- CCATTCAGCCAGATCGGCAT | ||||
| ISAba1 | F- ATGCAGCGCTTCTTTGCAGG | 393 | 50 | (24) |
| R- AATGATTGGTGACAATGAAG | ||||
| sul1 | F- CGGCGTGGGCTACCTGAACG | 433 | 58 | (21) |
| R- GCCGATCGCGTGAAGTTCCG | ||||
| sul2 | F- GCGCTCAAGGCAGATGGCATT | 293 | 58 | (21) |
| R- GCGTTTGATACCGGCACCCGT | ||||
| armA | F- ATTCTGCCTATCCTAATTGG | 315 | 56 | (21) |
| R- ACCTATACTTTATCGTCGTC | ||||
| PER-1 | F-ATGAATGTCATTATAAAAG | 920 | 45 | (25) |
| R-TTGGGCTTAGGGCAG |
Primers Used in This Study for Amplification of Genes in Acinetobacter baumannii Clinical Isolates
| Antibiotics | Drug Content | Zone of Inhibition, mm | Antibiotic Susceptibility Pattern | Resistance, % | ||
|---|---|---|---|---|---|---|
| S (>) | R (<) | S (>) | R (<) | |||
| Amikacin, µg | 30 | ≥ 17 | ≤ 14 | 65 | 25 | 27.27 |
| Ceftriaxone, µg | 30 | ≥ 21 | ≤ 13 | 00 | 90 | 100 |
| Ceftazidime, µg | 30 | ≥ 18 | ≤ 14 | 65 | 25 | 27.27 |
| Cefepime, µg | 30 | ≥ 18 | ≤ 14 | 33 | 57 | 63.33 |
| Ciprofloxacin, µg | 5 | ≥ 21 | ≤ 15 | 00 | 90 | 100 |
| Colistin, µg | 10 | MIC | 90 | 00 | 00 | |
| Co-trimoxazole, µg | 23.75 | ≥ 16 | ≤ 10 | 00 | 90 | 100 |
| Gentamicin, µg | 10 | ≥ 15 | ≤ 12 | 54 | 36 | 40.0 |
| Imipenem, µg | 10 | ≥ 16 | ≤ 13 | 70 | 20 | 22.2 |
| Meropenem, µg | 10 | ≥ 16 | ≤ 13 | 71 | 19 | 21.1 |
| Piperacillin-tazobactam, µg | 10 | ≥ 21 | ≤ 17 | 65 | 25 | 27.27 |
| Tigecycline, µg | 15 | ≥ 16 | ≤ 12 | 65 | 25 | 27.27 |
| Polymixin, units | 300 | MIC | 90 | 00 | 00 | |
| Tobramycin, µg | 10 | ≥ 15 | ≤ 12 | 90 | 00 | 00 |
| Tetracycline, µg | 30 | ≥ 15 | ≤ 11 | 33 | 57 | 63.33 |
Phenotypic Antimicrobial Sensitivity Pattern of Acinetobacter baumannii Clinical Isolates
3.4. Sequencing and Phylogenetic Analysis
The carbapenemase resistance genes, blaOXA-23 (extrinsic) and blaOXA-51 (intrinsic), were subjected to sequencing and phylogenetic analysis. Sequencing was performed using commercial sequencing services from Macrogen (Korea). We used NCBI-Nblast to determine the similarity index of the obtained sequences with already submitted sequences. The direct sequenced positive isolates were aligned to the reference sequences using phylogeny.fr software for phylogenetic tree construction.
3.5. Statistical Analysis
A chi-square test with SPSS version 21.0 software was used to determine the correlation between phenotypic and genotypic resistance patterns. A P-value of < 0.05 was considered significant.
4. Results
4.1. Characteristics of Specimens and Isolates
Out of 593 samples, 90 were found positive for A. baumannii. The gender-wise prevalence of A. baumannii isolates was 56 (62.2%) in males and 34 (37.8%) was in females. The prevalence of A. baumannii was 17.7% in wound samples (n = 16), 21.1% in blood samples (n = 19), 30% in pus samples (n = 27,) and 31.1% in burn samples (n = 28). The sites of infection were the respiratory tract of hospital intensive-care patients (n = 46, 51.1%), blood intensive-care patients (n = 20, 22.2%), urinary tract (n = 14, 15.5%), surgical soft tissue (n = 6, 6.6%), bone and joint (n = 2, 2.2%), and central nervous system lesion (n = 2, 2.2%).
4.2. Antimicrobial Susceptibility Pattern
The antibiotic resistance patterns against amikacin (AK), ceftriaxone (CRO), ceftazidime (CAZ), cefepime (FEP), ciprofloxacin (CIP), colistin (CT), co-trimoxazole (SXT), gentamicin (CN), imipenem (IPM), meropenem (MEM), piperacillin-tazobactam (TZP), tigecycline (TGC), polymixin-B (PB), tobramycin (TOB), and tetracycline (TE) are shown in Table 2. All isolates (100%) demonstrated resistance to a minimum of three classes of antibiotics and thus met the MDR criteria. In A. baumannii isolates, CT, PB, TOB, AK, CAZ, TZP, TGC, IPM, and MEM were most sensitive, while CIP, CRO, FEP, SXT, TE, and CN were most resistant. Multi-drug resistant A. baumannii was highly resistant to CIP, CRO, FEP, SXT, TE, AK, CAZ, and TE. A gradual increase in carbapenem resistance up to 20 (23%) was noted in the present bacterial isolates (Table 2).
4.3. Distribution of Antibiotic Resistance Genes
The PCR amplification of all the 13 resistance genes showed that the prevalence of blaOXA-23, blaOXA-51, blaOXA-40, blaNDM1, blaIMP, blaVIM, ISAba1, ssul1, sul2, armA, and PER-1 was 73% (63/90), 90% (81/90), 64.4% (58/90), 92.2% (83/90), 90% (81/90), 40% (36/90), 85.5% (77/90), 16.6% (15/90), 20% (18/90), 32.2% (29/90), and 12.2% (11/90), respectively. The blaOXA-24 and blaOXA-58 markers of class D carbapenemases were not detected. A p-value of < 0.05 was obtained for the relationship between genotypic and phenotypic resistance patterns of A. baumannii isolates. The overall statistical resistance rate between drugs and genes was 22.22% (20/90), and its prevalence was illustrated individually (Tables 3 and 4).
| Genes | Imipenem | Meropenem | ||
|---|---|---|---|---|
| Sensitive (N = 70) | Resistance (N = 20) | Sensitive (N = 71) | Resistance (N = 19) | |
| blaOXA-23 | ||||
| Positive (n = 63) | 49 | 14 | 60 | 11 |
| Negative | 21 | 06 | 11 | 08 |
| blaOXA-51 | ||||
| Positive (n = 81) | 61 | 20 | 62 | 19 |
| Negative | 09 | 00 | 09 | 00 |
| blaOXA-40 | ||||
| Positive (n = 58) | 40 | 18 | 42 | 16 |
| Negative | 30 | 02 | 29 | 03 |
| blaNDM1 | ||||
| Positive (n = 83) | 63 | 20 | 64 | 19 |
| Negative | 07 | 00 | 07 | 00 |
| blaIMP | ||||
| Positive (n = 81) | 61 | 20 | 62 | 19 |
| Negative | 09 | 00 | 09 | 00 |
| blaVIM | ||||
| Positive (n = 36) | 18 | 18 | 18 | 18 |
| Negative | 52 | 02 | 53 | 01 |
| ISAba1 | ||||
| Positive (n = 77) | 59 | 18 | 61 | 16 |
| Negative | 11 | 02 | 10 | 03 |
| sul1 | ||||
| Positive (n = 15) | 12 | 03 | 10 | 05 |
| Negative | 58 | 17 | 61 | 14 |
| sul2 | ||||
| Positive (n = 18) | 14 | 04 | 14 | 04 |
| Negative | 56 | 16 | 57 | 15 |
| armA | ||||
| Positive (n = 29) | 20 | 09 | 22 | 07 |
| Negative | 50 | 11 | 49 | 12 |
| PER-1 | ||||
| Positive (n = 11) | 10 | 01 | 10 | 00 |
| Negative | 60 | 19 | 61 | 19 |
Resistance Patterns of Imipenem and Meropenem According to the Individual Genes Among Clinical Isolates
| Sr. Numbers | Gene | Imipenem resistance rate | Meropenem resistance rate |
|---|---|---|---|
| 1 | blaOXA-23 | 22.22222222 | 17.46031746 |
| 2 | blaOXA-51 | 31.74603175 | 30.15873016 |
| 3 | blaOXA-40 | 28.57142857 | 25.3968254 |
| 4 | blaNDM1 | 31.74603175 | 30.15873016 |
| 5 | blaIMP | 31.74603175 | 30.15873016 |
| 6 | blaVIM | 28.57142857 | 28.57142857 |
| 7 | ISAba1 | 28.57142857 | 25.3968254 |
| 8 | sul1 | 4.761904762 | 7.936507937 |
| 9 | sul2 | 6.349206349 | 6.349206349 |
| 10 | armA | 14.28571429 | 11.11111111 |
| 11 | PER-1 | 1.587301587 | 0 |
Phenotypic and Genotypic Resistance Rate
4.4. Sequencing and Phylogenetic Analysis
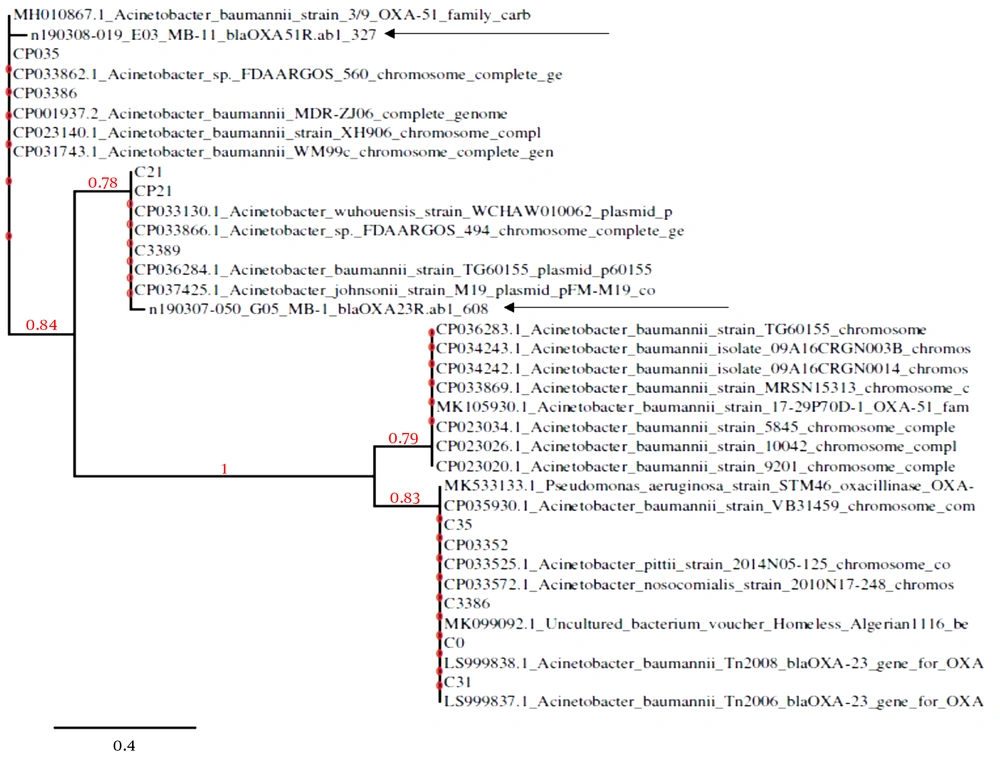
Different genotypes of CRAB isolates circulating in Pakistan, based on the NCBI data bank, were explained. The results disclosed that the isolates could be clustered into cardiographs, mostly represented by clade1 to clade4. The phylogenetic tree analysis indicated that blaOXA-51 was clustered in clade1 with its closely related species together. The strains were phylogenetically distinct from others and still not reported in Pakistani isolates. Similarly, blaOXA-23-like gene analysis was found in clade 2 after clade 1 and was reported for the first time in a Pakistani isolate. The relation of these resistance gene sequences with the closely related species represented 96% - 99% similarities to already submitted sequences in the NCBI databank. The phylogenetic tree represented the correlation of the strain with closely related species. The tree was generated following the neighbor-joining method (Figure 1 and Table 5).
| Gene | GenBank Accession Number | Closely Related Taxa Identified | Sequence Identity, % | Sequence Query Coverage, % |
|---|---|---|---|---|
| blaOXA-23 | LC096090.1 | Acinetobacter baumannii strain KKG5 | 98 | 25 |
| blaOXA-23 | LC096088.1 | A. baumannii strain KKG3 | 97 | 26 |
| blaOXA-23 | LC096087.1 | A. baumannii strain KKG2 | 98 | 25 |
| blaOXA-23 | LC096086.1 | A. baumannii strain KKG1 | 98 | 25 |
| blaOXA-51 | MH010867.1 | A. baumannii strain 3/9 OXA-51 | 95 | 84 |
| blaOXA-51 | CP036283.1 | A. baumannii strain TG60155 | 95 | 84 |
| blaOXA-51 | CP035930.1 | A. baumannii strain VB31459 | 95 | 84 |
Relationship Between Resistance Genes and Closely Related Taxa Described Using nBlasta
5. Discussion
For the past 60 years, β-lactam antibiotics have been amongst the most successful drugs used for the treatment of bacterial infections in humans. Acinetobacter has emerged as a significant class of pathogens, presenting continuous threats and challenges to the health care system throughout the world (20, 26, 27). The CRAB isolates created major therapeutic problems in the hospitals examined (1, 3, 28). In the current study, we investigated the occurrence of β-lactamase and carbapenemase-producing A. baumannii in patients admitted to the ICUs of a tertiary care hospital in Lahore, Pakistan. The results of the present study showed that of a total of 457 samples for bacterial culture, 90 (19.6%) were positive for A. baumannii, 82% for E. coli, 89% for Klebsiella (89%), and 63% for Pseudomonas spp. A similar study was conducted in Iraq on a total of 112 samples, in which most samples were positive for A. baumannii, while the other organisms were Candida albicans, Staphylococcus sp., P. aeruginosa, E. coli, and K. pneumoniae (29).
A previous study reported the appearance of carbapenem-resistant A. baumannii in Pakistan and showed increased resistance to cephalosporin, sulfamethoxazole, and beta-lactam antibiotics (30). They reported that the most sensitive antibiotics were tigecycline (80%) and colistin (50%). However, the results of the current study showed that CRAB strains were 100% resistant to CIP, CRO, and SXT. Tetracycline was found moderately effective against A. baumannii, indicated by the antibiograms and minimum inhibitory concentrations (MICs). Biglari et al. (31) reported that the isolates were most resistant to carbapenems and cephalosporin (70%) with high MIC values. Except for colistin, tetracycline, and rifampicin, the difference in resistance between the ICUs and other units was statistically significant (P < 0.05). Similar results were also reported in China (32) and Iraq (29). In the present study, the isolates were 100% resistant to ceftriaxone, ciprofloxacin, and co-trimoxazole, while moderate resistance was noted against gentamycin, piperacillin-tazobactam, and tigecycline. No resistance was noted against colistin, polymixin, and tobramycin. The reason behind the variations in antibiotic susceptibility patterns of A. baumannii could be due to the prolonged hospitalization because all the samples were collected from patients who were admitted to ICUs.
Carbapenemases represent the most versatile family of β-lactamases. These enzymes with catalytic efficiencies for carbapenem hydrolysis, resulting in elevated carbapenem MICs, include enzymes from classes A, B, and D (17). Investigations of the present study included genes from class B (blaIMP, blaNDM, and blaVIM), class D (blaOXA 23, blaOXA 24, blaOXA 40, blaOXA 51, and blaOXA 58), sulfonamide resistance genes (sul1 and sul2), aminoglycoside resistance methyltransferase gene (armA), an enzyme associated with blaOXA (ISAba1), and the PER1 gene. The most prevalent genes in the current study were blaNDM1 (92.2%), blaIMP (90%), blaOXA 51 (90%), ISAba1 (85%), blaOXA 23 (70%), and blaOXA 40 (64%). Besides, blaVIM was detected in 40% of total isolates, while the prevalence of armA, sul2, sul1, and PER1 was 32%, 20%, 16.6%, and 12%, respectively. In a previous study from Pakistan, the prevalence of the blaOXA-23 gene was 23.7% (26), 51.8% (14/27) in Switzerland (1), and 75.4% in Tehran (9).
A previous study from Iraq reported that genotypically identified A. baumannii represented resistance to all of the investigated β–lactam antibiotics. Besides, blaOXA-51, blaIMP, blaNDM, and blaOXA-23 were seen in 100%, 87.5%, 62.5%, and 59.4% of isolates (19). A similar study from Pakistan reported that in CRAB isolates, blaOXA-24, blaOXA-58, blaIMP, blaVIM, and blaSIM were completely absent (30). A similar result of blaOXA-24-like and blaOXA-58-like genes was also seen in the present study, as in the present study, neither of the genes was detected.
5.1. Conclusions
This study provides information about treating drug-resistant A. baumannii and the relationship of β-lactamases with the phenotypic resistance patterns. The co-existence of multiple drug-resistant bodies and virulent genes has important implications for the treatment of patients. The genotypic resistance pattern was closely related to the phenotypic patterns by detecting the resistance genes using PCR and antimicrobial susceptibility testing by disk diffusion method. This study provides information about treating the drug-resistant A. baumannii and also the relationship of virulent genes with phenotypic resistance patterns.