1. Background
The human gut microbiome contains members of all domains of life and plays an essential role in health and disease (1). However, its fungal component has received little attention compared to bacteria. In recent years, next-generation sequencing (NGS) techniques, targeting either ribosomal DNA (rDNA) or internal transcribed spacer (ITS) regions, have been recorded as reliable methods in the identification of the whole fungi populations in the gastrointestinal (GI) tract. However, despite their strengths, NGS methods are not able to distinguish between transient and resident gut flora. In this context, culture-based methods remain useful to display the viable culturable portion of fungi, shed light on numerous aspects, such as species interaction and the contribution of diet and environment to the gut fungal composition, and allow functional analyses such as antifungal susceptibility (2).
Members of Candida genus are the most commonly detected fungi in mycobiome studies (3, 4). However, among this genus, only few species could be found as natural, asymptomatic components of the human mycobiome, and consequently, may play a role in gut ecology or host health (5). Several of these human resident yeasts, such as Candida albicans, C. glabrata, C. tropicalis, and C. parapsilosis, use the mammalian digestive tract, the skin, and the mucosal surfaces as primary niches (4). Their colonization starts at birth, continues during life, and is influenced by multiple factors, including host, trans-kingdom interactions between fungi and bacteria, diet, and lifestyle (6, 7). Other Candida species, such as C. krusei, are considered as passengers or potential colonizers from dietary and environmental sources (8). However, as described for other yeasts of non-human origin (like Saccharomyces), they probably interact with other yeast species and play a role in gut homeostasis (3).
In some conditions, Candida species are able to switch from commensal to pathogen forms causing superficial or invasive infections. This capacity is provided by the expression of virulence factors, such as biofilm formation and factors of fitness in the face of stresses. Invasive candidiasis (IC), which affects more than 250,000 people and causes more than 50,000 deaths worldwide every year (9), is mainly caused by C. albicans. It is chiefly a result of the widespread use of antifungals. Antifungal resistance, mainly against fluconazole, has emerged worldwide, and the proportion of isolated C. albicans from IC has decreased in favor of non-albicans Candida (NAC) resistant species (10).
2. Objectives
Our aim was to phenotypically and genotypically characterize culturable Candida isolates present in stool specimens of healthy Tunisian individuals and evaluate their antifungal susceptibility.
3. Methods
3.1. Yeast Isolation
Yeasts were isolated from 46 stool specimens provided by the Laboratory of Parasitology-Mycology, Pasteur Institute of Tunis. Fecal samples were self-collected within the last 24 hours by healthy Tunisian adults in the setting of routine diagnosis. Stool specimens were spread-plated on Sabouraud dextrose agar (SDA) supplemented with chloramphenicol (100 µg/mL), and then incubated at 37°C for 48 hours. Isolates growing in pure culture were identified at species and genomic levels and tested for phenotypical features to resist oxidative stress, acidic conditions, antifungal drugs, and ability to form biofilm.
3.2. Species Identification
Conventional methods: For Candida species identification, all yellowish cream colonies evocating Candida genus were subcultured at 37°C for 48 hours on chromogenic medium (CHROMagarTMCandida, Media Pioneer, Germany). Yeast isolates not identified by CHROMaga were analyzed using the API ID32C (Biomerieux, France) according to the manufacturer’s instructions. ITS PCR-sequencing: DNA was extracted from yeast isolates using the QIAamp DNA Mini kit (Qiagen, Germany). The 18S rRNA gene partial sequence, ITS region complete sequence, and 28S rRNA gene partial sequence were amplified and sequenced using fungal primers ITS5 (forward): (5’-GGAAGTAAAAGTCGTAACAAGG-3’) and ITS4 (reverse): (5’-TCCTCCGCTTATTGATATGC-3’) (11). Nucleotide sequencing was carried out in the Applied Biosystems™ 3500 sequencer calibrated with BigDye® Terminator V3.1. DNA sequences were assembled and edited using BioEdit Sequence Alignment Editor software version 7.0.5.3. The consensus sequences obtained were compared to sequences in the Basic Local Alignment Search tool (BLAST). The sequences generated in this study were deposited in GenBank under the following accession numbers: MT777621-MT777676 (Appendix 1 in Supplementary File).
3.3. Phenotypic Characterization of Candida Isolates
All tests were performed in yeast extract peptone dextrose (YPD) medium and in duplicate. Isolate resistance to the different stresses was assessed by counting colony-forming units (CFU) on YPD agar plates.
3.3.1. Resistance to Oxidative Stress
Candida suspension (~106 cells/mL) was grown for 2 hours in liquid medium at 37°C in the presence of hydrogen peroxide (H2O2) in different concentrations (i.e., 0.5 mM, 2 mM, 4 mM, and 8 mM). A YPD medium without H2O2 was used as control. Then, 100 µL of the diluted culture (1:100) was plated and incubated at 30°C until the appearance of colonies. The percentage of survival was calculated after counting the number of colonies on each plate (12).
3.3.2. Resistance to Acidic Conditions
Candida suspension (~105 cells/mL) was grown overnight in liquid media (pH2 and pH3) at 37°C. Yeast extract peptone dextrose medium (pH = 6.5) was used as control. The following steps were performed as described in the oxidative test.
3.3.3. Biofilm Test
Biofilm test was performed in triplicate on YPD according to Sciavilla et al. (13) and was quantified by optical density measurement at 570 nm with a microplate reader (Synergy2BioTek, Winooski, VT, USA).
3.4. Antifungal Susceptibility Testing
Candida isolates were tested for susceptibility to six antifungal drugs (i.e., fluconazole, voriconazole, caspofungin, micafungin, amphotericin B, and 5-flucytosine) by the automated VITEK 2 compact system (BIoMerieux, France) using AST-YS08 cards and following manufacturer’s instructions. The minimum inhibitory concentration (MIC) was established, and the results interpretation was performed using the breakpoints defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute (CLSI) (14, 15).
3.5. Intra- and Interspecific Diversities Analysis of Candida Species ITS Sequences
The ITS1-5.8S-ITS2 of each Candida species was aligned to the reference/type strain in megaX software version 7.0. Intraspecific variability was performed using DnaSP version 6.12.03., considering gaps. The analysis was based on the number of haplotypes (Hap), variable sites, haplotype diversity (Hd), and nucleotide diversity (Pi). Overall, 27 Candida strains (isolated from human feces) from GenBank and eight reference/type Candida strains presenting the longest sequence (Appendix 2 in Supplementary File) were considered for the construction of a maximum likelihood analysis performed by MegaX and based on the GTR model (G + I).
3.6. Statistical Analysis
Wilcoxon rank-sum test was used to analyze the ability of Candida isolates to grow under different stressful conditions, and t-test method was run to compare the capacity of Candida isolates to form biofilm. Statistical analysis was performed-using “rstatix” R package (version 0.7.0) and “ggpubr” R package (version 0.4.0), respectively. Boxplots were built using the “ggplot2” R package (version 3.3.3).
4. Results
4.1. Yeast Isolation
Among the 46 stool specimens subjected to Sabouraud culture, 37 (80%) showed the presence of yeasts. A total of 56 yeast isolates were recorded from positive fecal specimens and submitted to species identification. The recorded Candida isolates were subjected to phylogenetic analysis and phenotypic characterization.
4.2. Species Identification
The number of isolates, according to species, is reported in Table 1. Species identification results using conventional methods (CHROMagarTMCandida + ID32C) and PCR sequencing were concordant for 55 isolates out of 56 (98%). C. albicans was the most isolated species (n = 21), which was present in about 56.7% of positive samples (Table 1). It was isolated alone in 14 samples, and in the remaining cases, it was associated with the three most dominant species, namely C. parapsilosis (27%), C. glabrata (24.3%), and C. krusei (24.3%) (Table 1).
| Species | Species Identification Method | Percentage***, % | |
|---|---|---|---|
| ITS Sequencing* | Conventional Methods** | ||
| Candida albicans | 21 | 21 | 56.7 |
| C. dubliniensis | 1 | - | 2.7 |
| C. tropicalis | 1 | 1 | 2.7 |
| C. parapsilosis | 10 | 9 | 27 |
| C. glabrata | 9 | 9 | 24.3 |
| C. kefyr | 1 | 1 | 2.7 |
| C. krusei | 9 | 9 | 24.3 |
| Geotrichum capitatum | 2 | 2 | 5.4 |
| Rhodotorula mucilaginosa | 1 | 1 | 2.7 |
| Trichosporon asahii | 1 | 1 | 2.7 |
| Total isolates | 56 | 55 | |
a*, Reference test, **, only concordant results with ITS-sequencing were considered; ***, percentage calculated on the base of the number of stool specimens with yeasts (n = 37).
4.3. Candida Phenotypic Analysis
4.3.1. Oxidative Stress, Extreme pH, and Biofilm Formation
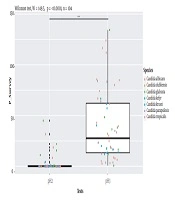
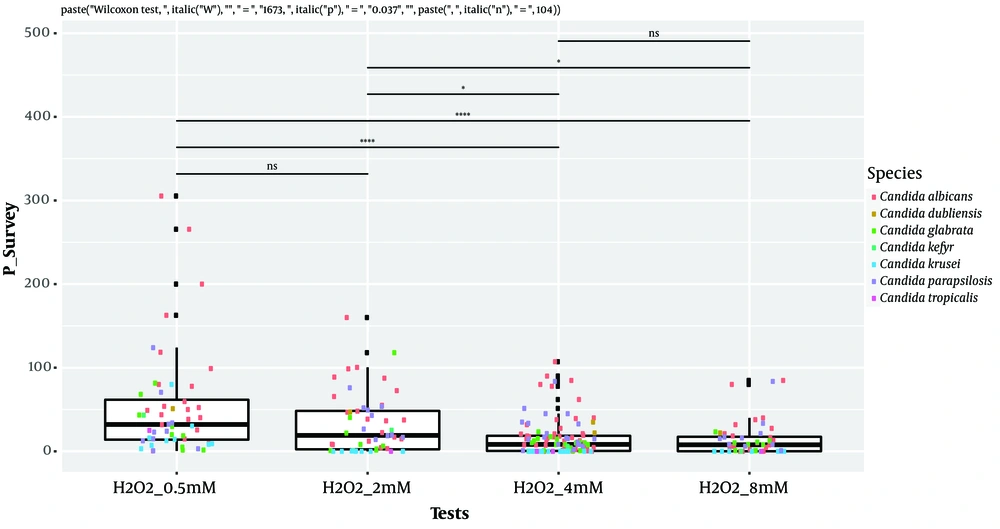
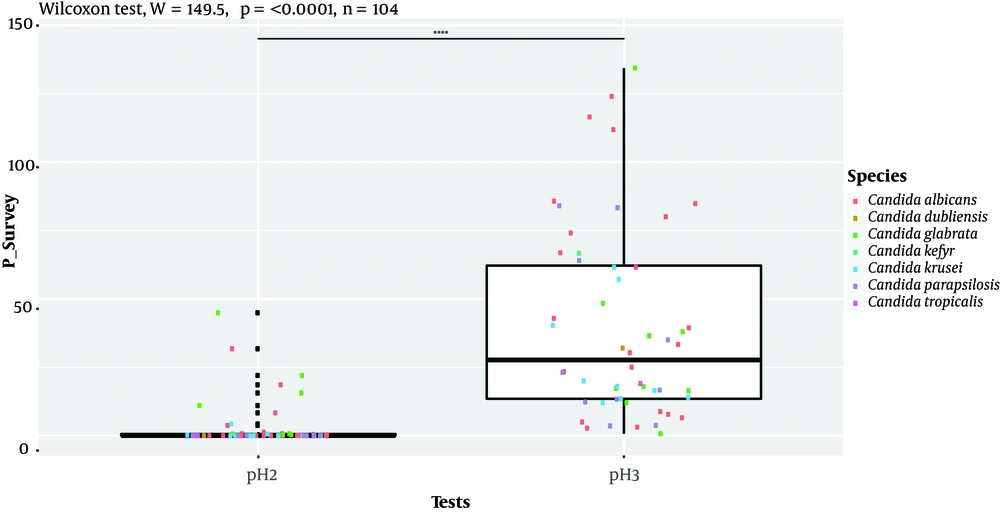
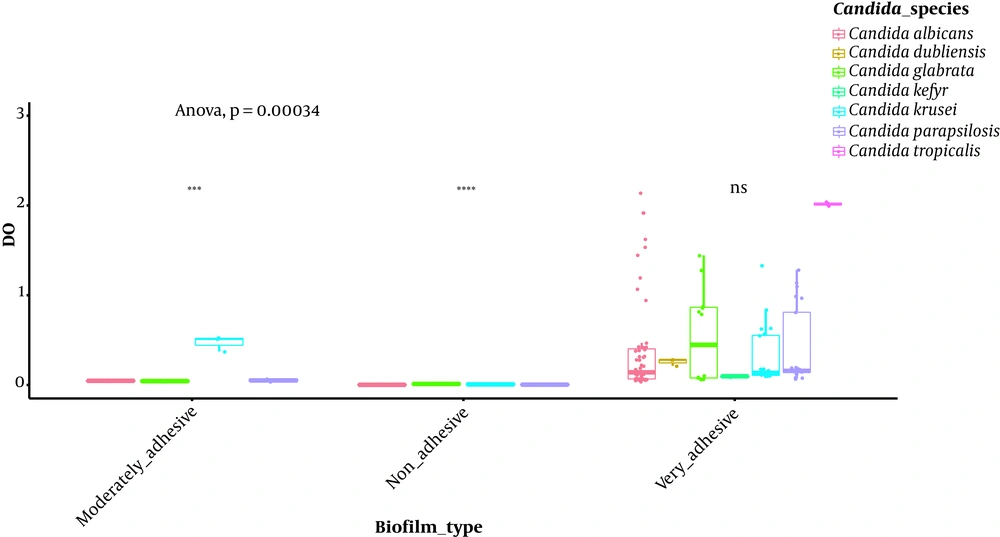
Isolates’ ability to resist under some gastrointestinal tract (GI) conditions is reported in Appendix 3 in Supplementary File. The statistical analysis revealed a significant difference between tests performed at 0.5 Mm H2O2 versus 4 mM H2O2 and 0.5 Mm versus 8 mM H2O2 (P < 0.0001; Figure 1). In general, 96.15% of the isolates are able to grow at 0.5 mM, while their survival decreased under 2 mM (76.92%), 4 mM (75%), and 8 mM (63%5) conditions. A strongly significant difference was observed between pH tests (P < 0.0001; Figure 2). Here, 98% and 19% of the isolates were able to grow at pH = 3 and pH = 2, respectively. Among the 52 isolates, 46 showed the ability to form biofilm. The positive ones were divided into very adhesive (86.95%) and moderately adhesive (13.04%) isolates. A significant difference between the two groups was recorded (Figure 3).
4.4. Susceptibility of Candida Species Against Antifungal Drugs
Isolate susceptibility to the six antifungal drugs is shown in Table 2 and Appendix 3 in Supplementary File. The results revealed that only C. albicans were resistant to fluconazole (MIC: 8 - 16 µg/mL) except C. krusei, which has an intrinsic resistance to this antifungal. Fluconazole presented the highest number of resistant isolate (23.8% of C. albicans), whereas only 22.2% of C. glabrata and 11.1% of C. albicans were resistant to caspofungin (MIC: 0.5 - 1 µg/mL), in addition one C. glabrata isolate was resistant to micafungin (MIC: 1 µg/mL). Finally, no resistance was shown against voriconazole, amphotericin B and 5-flucytosine.
| Minimum Inhibitory Concentration (MIC) Range, µg/mL | ||||||
|---|---|---|---|---|---|---|
| Fluconazole | Voriconazole | Caspofungin | Micafungin | Amphotericin B | 5-Flucytosine | |
| Candida albicans (n = 21) | ≤ 0.5 - 16 | ≤ 0,12 | ≤ 0.12 - 1 | ≤ 0.06 - 0,5 | ≤ 0.25 - 1 | ≤ 1 |
| C. glabrata (n = 9) | ≤ 0.5 - 8 | ≤ 0.12 - 0.25 | ≤ 0.2 - 1 | ≤ 0.6 - 0.5 | 0.5 - 1 | |
| C. parapsilosis (n = 10) | ≤ 0.5 - 1 | ≤ 0.12 | 0.25 - 1 | 0.12 - 2 | ≤ 0.5 - 1 | |
| C. krusei (n = 9) | 8 - 16 | ≤ 0.12 | 0.5 | 0.12 | 0.5 - 1 | 8 - 16 |
| C. tropicalis (n = 1) | 1 | ≤ 0.12 | ≤ 0.12 | ≤ 0.5 | ≤ 0.5 | ≤ 1 |
| C. dubliniensis (n = 1) | 1 | ≤ 0.12 | 0.25 | ≤ 0.06 | 0.5 | |
| C. kefyr (n = 1) | ≤ 0.5 | ≤ 0.12 | ≤ 0.12 | 0.12 | 0.5 | |
4.5. Intra- and Interspecific Variability of the ITS1-5.8S-ITS2 Region in Candida Species
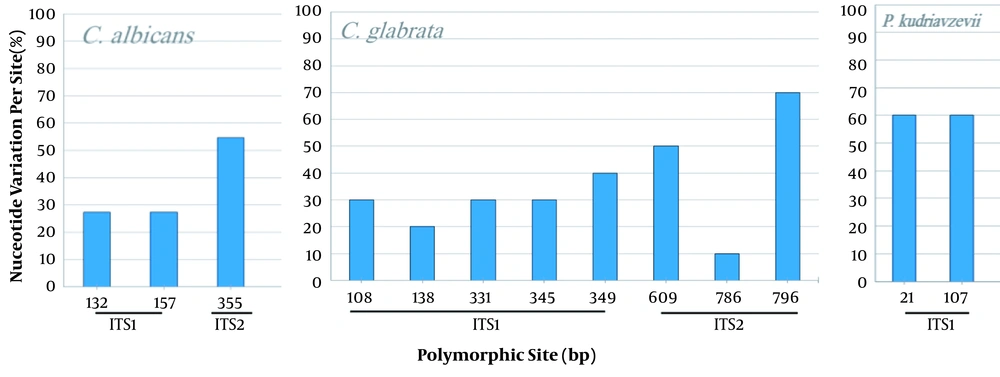
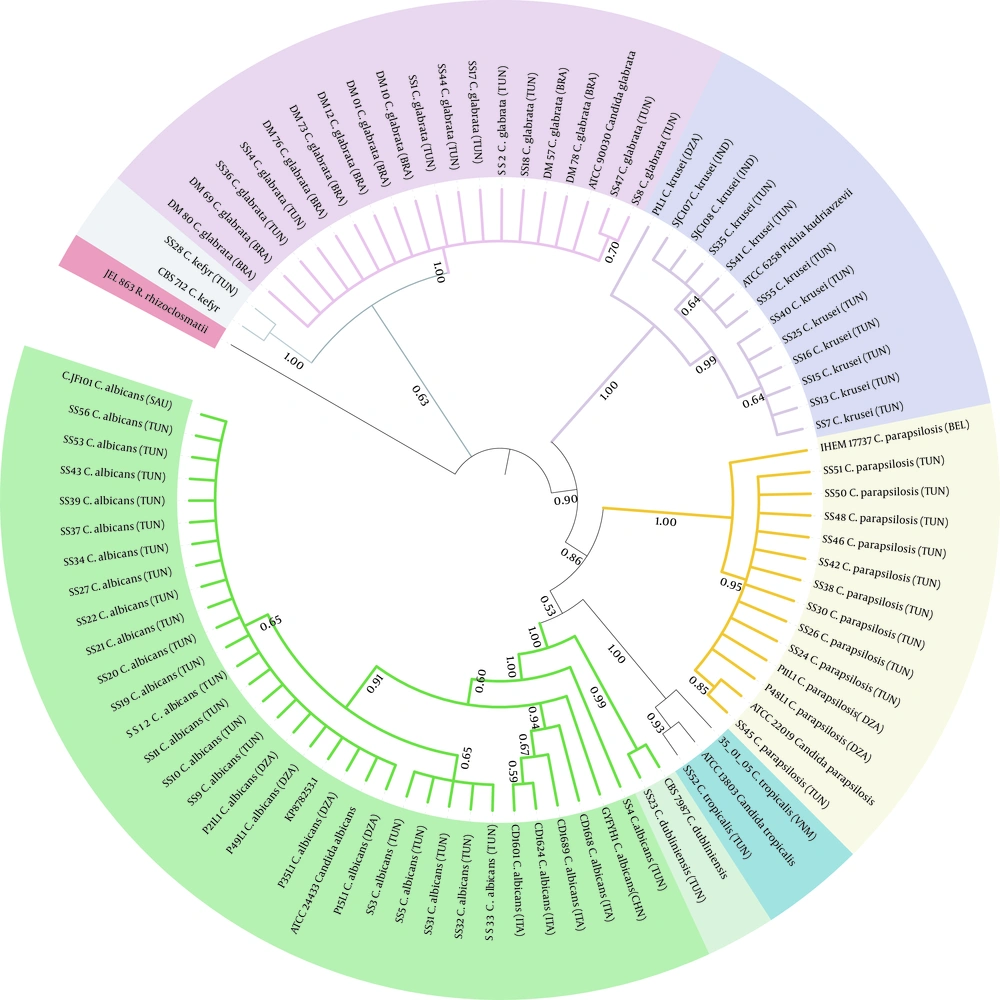
The comparison of the Candida spp. ITS sequences to the type/reference strains revealed the presence of a high intraspecific variability reflected by the numbers of haplotype diversity (Hd = 0.533 to 1) and nucleotide diversity (Pi = 0.002 to 0.01094) (Table 3). However, non-intraspecific variations were observed among C. parapsilosis isolates (sensu stricto). Candida species presenting haplotype variability and more than 10 stains were assessed for the polymorphism of the ITS region (Figure 4). Variable sites were mostly recorded in ITS1 than in ITS2, and no variability was shown in the 5.8S region. The phylogenetic tree was in agreement with the molecular identification of the Candida strains, which were consistently separated in clades at the species level (C. krusei clade: 3 groups; C. glabrata clade: 2 groups; C. albicans clade: 2 groups and 2 subgroups; C. parapsilosis clade: 1 group) (Figure 5).
| Candida species | Number of Sequences | Sequence Length, bp | Number of Haplotypes (Hap) | Haplotype Diversity (Hd) | Number of Variable Sites | Nucleotide Diversity (Pi) |
|---|---|---|---|---|---|---|
| Canddia albicans | 22 | 457 | 5 | 0.6753 | 3 | 0.002 |
| C. dubliniensis | 2 | 459 | 2 | 1 | 5 | 0.01094 |
| C. glabrata | 10 | 798 | 5 | 0.8667 | 8 | 0.00374 |
| C. parapsilosis (sensu stricto) | 11 | 449 | 1 | 0 | 0 | 0 |
| C. tropicalis | 2 | 470 | 2 | 1 | 1 | 0.00213 |
| Pichia kudriavzeviia | 10 | 471 | 2 | 0,533 | 2 | 0,00113 |
| Kluyveromycets marxianusa | 2 | 664 | 1 | 0 | 0 | 0 |
aPichia kudriavzevii = teleomorph of Candida krusei, Kluyveromycetes marxianus = teleomorph of Candida kefyr
Variability of the ITS1-5.8S-ITS2 region in Candida albicans, C. glabrata, and P. kudriavzevii. Nucleotide variation is performed by comparison to the related reference strain. C. albicans (532 pb, 22 sequences), C. glabrata, (877 pb, 10 sequences), and P. kudriavzevii (teleomorph of C. krusei), (470 pb, 10 sequences).
Phylogenetic relationships among Candida strains. A Neighbor-joining tree using Maximum Likelihood algorithm with 1000 bootstrap replicates was performed. Phylogenetic tree was generated using iTOL version 6. Strains from our study are reported using their code (SS1 to SS56). The 26 Candida strains selected from GenBank and Rozella rhizoclosmatii JEL 863 used as an outgroup are in bold. Country origins of each strain are indicated in parentheses and are designed according to the standard abbreviation: BRA, Brazil; CAN, Canada; CHI, China; DZA, Algeria; IND, India; ITA, Italy; SAU, Saudi Arabia; TUN, Tunisia; VNM, Vietnam; BEL, Belguim
5. Discussion
Our study focused on the phenotypic and genotypic characterization of cultivable yeasts isolated from stool specimens of Tunisian individuals. The yeast isolates were successfully identified by conventional methods in 96% of cases, which showed that Candida species was the predominant type (52 out of 56 isolates). These results corroborate the findings of other works studying the fungal populations in healthy human GI tract, which showed the scarcity of non-Candida genus (e.g., G. capitatum, T. asahii, and R. mucilaginosa) in stool (4). The controversial absence of Saccharomyces cerevisiae (3) might be associated with the limits of the culture-dependent methods (4) and/or dietary habits. Among the seven isolated Candida species, C. albicans was the most frequent, followed by C. parapsilosis and C. glabrata. The prevalence of these species in healthy adults’ stool specimens was already reported (3, 4) and was explained by the early colonization of the mucosal surfaces by means of mother-infant transmission (16). The fourth abundant species herein was C. krusei. It has been essentially isolated from the environment, and its presence in stool seems to be associated with diet (8).
Our set of isolates showed a capacity to grow under oxidative stress (0.5 mM) and acidic conditions (pH = 3), which is in concordance with a previous study findings (6). In addition, we found that C. albicans, C. glabrata, and C. parapsilosis were able to tolerate and persist under high H2O2 concentration, while C. krusei was not. This could confirm that having more fitness, these species are a part of true colonizers, whereas C. krusei is associated with transient flora. No significant difference was noted between tests performed at 2 - 8 mM, and this result should be taken into account for further investigations. A significant difference was found between pH = 2 and pH = 3 tests, and only a few strains were able to resist at pH = 2. This could suggest that Candida species are able to grow but not persist in extreme acidic conditions, which is in line with a previous microbiota ecology study (17). However, a shorter test incubation may shed light on the strain performance to colonize the human gut. As expected, all Candida species were able to form biofilm, and no significant difference was shown between the positive isolates. Hence, performing tests with mixed species in concordance with the associations found in our results (data not shown) would be pertinent.
The in vitro antifungal susceptibility study revealed that apart from C. krusei, which is intrinsically resistant to fluconazole, 33.3% of C. albicans isolates were resistant to fluconazole. The recent increase in antifungal resistance among human opportunistic fungi has been due to their frequent use of candidiasis treatments (10) and environmental exposure. Indeed, antifungals, especially azoles, are extensively used in plant protection and infection treatments in Tunisia, leading to the contaminated watersheds and lagoon ecosystems (18). Besides, the intermediate resistance against micafungin and caspofungin could be a prelude to the emergence of other types of resistance.
The ITS1-5.8S-ITS2 rDNA region, which constitutes a universal fungal barcode sequence (19), was used to study the genetic diversity. With the exception of C. parapsilosis, the ITS polymorphism investigation revealed a high intraspecific variability essentially in the ITS1 region, which concords with previous results (20). The fact that C. albicans and C. glabrata species are described as a part of intestinal and mucosal flora and the high intraspecific variation observed in most species could be a genetic adaptation to the environment (20). Yet, in our collection and in a previous report (20), C. albicans was two times more represented than C. glabrata and had two times less number of variable sites. This could be explained by the haploid state of C. glabrata, which relies on its high mutagenic potential to generate genetic diversity (21). In addition, the exogenous source of C. parapsilosis and its clonal mode of reproduction might explain the lack of genetic divergence observed. Finally, it is imperative to admit that the ITS region does not allow a good discrimination at the intraspecific level. Hence, with a better genetic marker, such as the multilocus sequence typing (MLST) or the random amplification of polymorphic DNA (RAPD), we will gain a better understanding of genetic variability (22).
5.1. Conclusions
In sum, this study is the first Tunisian investigation on intestinal Candida spp. in healthy individuals and englobing identification, phenotypic characterization, and antifungal susceptibility. It was based on culture-dependent methods, which allowed the identification of viable culturable Candida isolates. This method remains useful to better understand the intestinal fungal community, evaluate its ability to survive stressful conditions, express virulence factors, and resist antifungal drugs. Nevertheless, further complementary studies using culture-independent methods and NGS technology are needed for a better characterization of the gut mycobiome in Tunisia.