1. Background
Acinetobacter baumannii is an aerobic Gram-negative bacillus. This major pathogen causes various types of hospital-acquired infections, such as respiratory, urinary tract, and wound infections (1). Acinetobacter baumannii is resistant to major antibiotics, including β-lactams, aminoglycosides, and quinolones. Outbreaks of multidrug-resistant (MDR) bacteria significantly affect the treatment of patients with severe infections (2). Ciprofloxacin, levofloxacin, and sparfloxacin are representative quinolone antibiotics, which exhibit antimicrobial activity against Gram-positive and Gram-negative bacteria and have shown clinical efficacy in treating respiratory infections. They can be orally administered due to their stable chemical structures and excellent tissue permeability. Quinolones are known to exhibit excellent permeability as they usually enter the cells via porin (membrane transport protein) located in the outer membrane of bacterial cells (3). In bacteria, efflux pumps actively transport antibiotics from cells and are known to be the primary cellular components mediating the natural resistance of Gram-negative bacteria to various antibiotics (4).
In recent times, pulsed-field gel electrophoresis and multilocus sequence typing (MLST) have been used to trace the lineage or origin of most epidemic strains, including those of A. baumannii. Among the structural genes with varying sequences in different species, primers for gltA, gyrB, gdhB, recA, cpn60, gpi, rpoD, pgm, quiA, and pcaf are used for A. baumannii. Among these, the high allelic number of gyrB indicates frequent mutations, which also leads to the expression of various STs (5, 6). Multiplex PCR analysis using the gyrB primer was confirmed to be the most rapid method for distinguishing and identifying A. baumannii and four other A. baumannii complexes (5, 6). Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (MS) analysis identifies the measurable molecular weight (100 Da to 100 kDa) and the bacterial cell count corresponding to one or two colonies (105 to 106) by examining the protein peaks (7).
2. Objectives
The effluent pump inhibition influence test and sequencing of quinolone resistance-determining region (QRDR) of gyrA and parC genes were conducted to determine the cause of resistance of quinolone-based antibacterial agents. In addition, MLST was performed to determine the genetic correlation of the structural gene gyrB with the amino acid variation at a specific site of gyrA called QRDR. The protein molecular weight of each strain was measured and compared with the results of antimicrobial sensitivity tests.
3. Methods
3.1. Strain Isolation
This study was approved by the Institutional Review Board (IRB-202010027) of Dankook University. Ninety-eight strains of A. baumannii isolated from sputum, wound debris, urine, and pus samples obtained from the Gyeonggi Southern General Hospital from 2017 – 2019 were identified using the VITEK® 2 XL (bioMérieux, Marcy-l'Etoile, France) GN-card. The detection primers used in PCR included OXA-51-like F: 5'-TAA TGC TTT GAT CGG CCT TG-3' and OXA-51-like R: 5'-TGG ATT GCA CTT CAT CTT GG-3'. The PCR product of 353 bp was confirmed at 302 nm using the Molecular Imager Gel DOCTM XR+ System (Bio-Rad, USA) (8).
3.2. Antimicrobial Susceptibility Test
Antimicrobial susceptibility was assessed using the VITEK® 2 XL AST-N225 card for measuring the minimum inhibitory concentration (MIC). The following 17 antibiotics were used: penicillin with extended spectrum (piperacillin), β-lactam/β-lactamase inhibitor combinations (ampicillin/sulbactam, piperacillin/tazobactam, and ticarcillin/clavulanic acid), cephems (cefotaxime, ceftazidime, and cefepime), carbapenems (imipenem and meropenem), aminoglycosides (amikacin and gentamicin), folate pathway inhibitors (trimethoprim/sulfamethoxazole), quinolone (levofloxacin and ciprofloxacin), tetracyclines (minocycline), glycylcyclines (tigecycline), polymyxin (colistin), and monobactam (aztreonam). The antimicrobial susceptibility test was performed in compliance with the Clinical and Laboratory Standards Institute (CLSI) guidelines (2020, M100-S30) (9). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as standard strains.
3.3. Efflux Pump Inhibition Test
The efflux pump inhibition test was performed using phenylalanine-arginine β-naphthylamide (PAβN), an efflux pump inhibitor (EPI), on 50 strains with confirmed ciprofloxacin resistance in the antimicrobial susceptibility test. The second- and third-generation quinolone antibiotics, ciprofloxacin and levofloxacin (Sigma-Aldrich, USA) respectively, were used. PaβN was administered at 100 μg/mL, and comparison of MIC reduction of the antibiotic in presence and absence of EPI was measured (10). The MIC of the antibiotic was measured using the solid medium dilution method, as directed in the CLSI guidelines (2020, M100-S30) (9). E. coli ATCC 25922 was used as the standard strain.
3.4. Analysis of QRDR Sequence of gyrA and parC
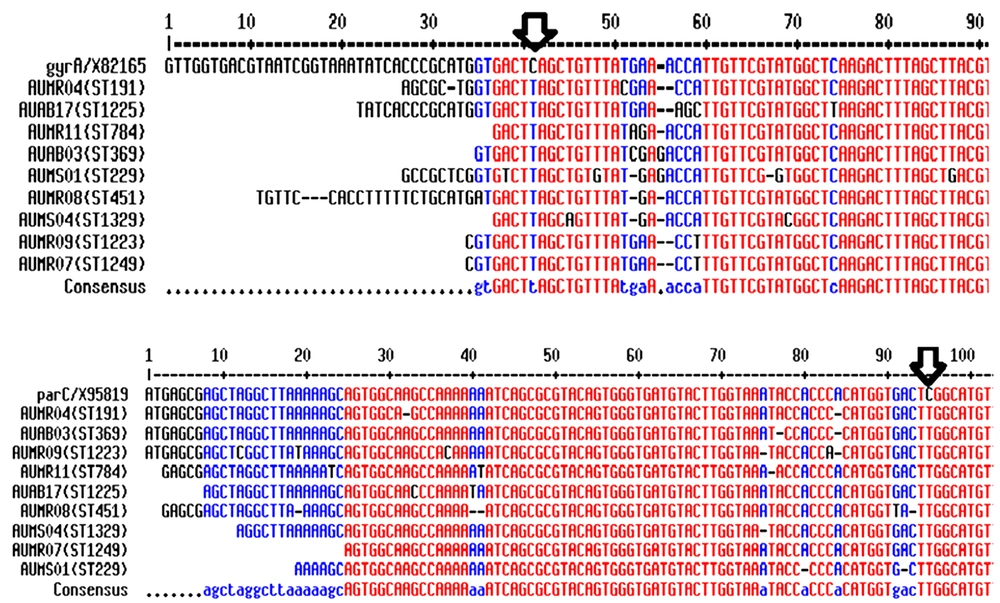
Sequence analysis was performed on 50 ciprofloxacin-resistant strains of A. baumannii to determine the presence of mutation in each gene (11). The analyzed nucleotide sequences were the standard A. baumannii gyrA and parC sequences. The gyrA and parC sequences of the strains were compared to the corresponding standard gene sequences (GenBank accession numbers X82165 and X95819, respectively) (12).
3.5. Multilocus Sequence Typing Analysis
Multilocus sequence typing analysis was performed as directed in PubMLST (13). Information regarding the seven structural genes used in the study was confirmed from the MLST website and used for primer designing, followed by PCR (14). Each of the seven structural genes was provided a unique allelic number through PubMLST, and the final ST of A. baumannii was determined using a combination of the unique alleles (Table 1). The correlation between the allelic number of gyrB, a structural gene of A. baumannii, and STs was investigated. The relationship between the strains was confirmed by comparing the results of the antimicrobial susceptibility test and a peak analysis (performed using the MALDI-TOF MS).
| Locus | Primer Sequence (5 - 3) | Amplicon Size (bp) |
|---|---|---|
| gltA | 722 | |
| Citrato F1: | AATTTACAGTGGCACATTAGGTCCC | |
| Citrato R12: | GCAGAGATACCAGCAGAGATACACG | |
| gyrB | 594 | |
| gyrB_F: | TGAAGGCGGCTTATCTGAGT | |
| gyrB_R: | GCTGGGTCTTTTTCCTGACA | |
| gdhB | 774 | |
| GDH SEC F: | ACCACATGCTTTGTTATG | |
| GDH SEC R: | GTTGGCGTATGTTGTGC | |
| recA | 425 | |
| RA1 | CCTGAATCTTCYGGTAAAAC | |
| RA2 | GTTTCTGGGCTGCCAAACATTAC | |
| cpn60 | 640 | |
| cpn60_F: | GGTGCTCAACTTGTTCGTGA | |
| cpn60_R: | CACCGAAACCAGGAGCTTTA | |
| gpi | 456 | |
| gpi_F: | GAAATTTCCGGAGCTCACAA | |
| gpi_R: | TCAGGAGCAATACCCCACTC | |
| rpoD | 672 | |
| rpoD_F: | ACCCGTGAAGGTGAAATCAG | |
| rpoD_R: | TTCAGCTGGAGCTTTAGCAAT |
Oligonucleotides Used for Detecting Housekeeping Genes
3.6. MALDI-TOF MS Analysis
MALDI-TOF MS analyzed the differences in the protein composition of A. baumannii and distinguished between the different groups of Acinetobacter. This method confirmed the spectral peak of a ciprofloxacin-resistant A. baumannii strain. A detailed analysis was performed on the spectrum obtained for the A. baumannii target strain in the region between 2,000 m/z and 18,000 m/z using the SARAMIS database (15).
4. Results
4.1. Strain Isolation
Of the 98 strains of A. baumannii isolated from clinical specimens, 64 were eventually selected through biochemical testing using VITEK® 2 XL (bioMérieux) and PCR detection of the blaoxa-51-like gene.
4.2. Antimicrobial Susceptibility Test
The resistance rates of A. baumannii were 78.1% (n = 50) for imipenem and meropenem, 100% (n = 64) for aztreonam, 79.7% (n = 51) for cefotaxime, 73.4% (n = 47) for ceftazidime, 79.7% (n = 51) for cefepime, 45.3% (n = 29) for aminoglycosides, and 59.3% (n = 38) for amikacin and gentamicin. Among the quinolones tested, a resistance rate of 78.1% (n = 50) was observed for ciprofloxacin. Meanwhile, multidrug-resistant A. baumannii (MRAB), which is simultaneously resistant to at least three antibiotic classes (including β-lactams, carbapenems, aminoglycosides, and quinolones), showed 57.8% resistance (n = 37).
4.3. Efflux Pump Inhibition Test
Fifty ciprofloxacin-resistant strains (MIC ≥ 4 μg/mL, as indicated in the antimicrobial susceptibility test) were evaluated in the efflux pump inhibition test using PaβN (Appendix 1 in Supplementary File). The MIC of ciprofloxacin for 18 strains was lower in presence of PaβN than that in its absence, and the reduction in MIC was four-fold for 2 strains and two-fold for 16 strains. The MIC of levofloxacin reduced for 43 strains. The reduction was eight-fold for 2 strains, four-fold for 18 strains, and two-fold for 23 strains.
4.4. Sequence Analysis of QRDR in gyrA and parC
In all 50 strains of A. baumannii, the codon for serine (TCA) at the 83rd position in the gyrA sequence was replaced by that for leucine (TTA) by a point mutation. In 43 strains, the codon for serine (TCG) at the 80th position in parC sequence was replaced by that for leucine (TTG); this mutation was not observed in 7 strains. Therefore, mutations were identified in both gyrA and parC in 43 strains (Figure. 1).
4.5. Multilocus Sequence Typing Analysis
Multilocus sequence typing analysis of 64 A. baumannii strains showed 18 STs and allelic numbers that determined the STs (Table 2). The most frequently detected allele expressed in five types of STs, including ST191, and was gyrB3 (62.5%; n = 40). ST191, ST229, ST369, ST451, ST784, ST1223, ST1225, ST1249, and ST1329 showed resistance to ciprofloxacin, whereas ST229, ST369, ST451, ST784, ST1223, ST1225, ST1249, and ST1329 showed resistance to imipenem, meropenem, gentamicin, and amikacin. ST191 showed resistance to ciprofloxacin, imipenem, meropenem, amikacin, and gentamicin in 21 strains and intermediate resistance in 1 strain (Table 3).
| ST | Number of Isolates (%) | Allelic Number | ||||||
|---|---|---|---|---|---|---|---|---|
| gltA | gyrB | gdhB | recA | cpn60 | gpi | rpoD | ||
| ST191 | 25 (39.0) | 1 | 3 | 3 | 2 | 2 | 94 | 3 |
| ST195 | 2 (3.1) | 1 | 3 | 3 | 2 | 2 | 96 | 3 |
| ST229 | 6 (9.3) | 1 | 15 | 2 | 28 | 1 | 107 | 32 |
| ST369 | 3 (4.6) | 1 | 3 | 3 | 2 | 2 | 106 | 3 |
| ST451 | 5 (7.8) | 1 | 3 | 3 | 2 | 2 | 142 | 3 |
| ST552 | 2 (3.1) | 21 | 35 | 2 | 28 | 1 | 145 | 4 |
| ST595 | 1 (1.5) | 1 | 15 | 66 | 12 | 33 | 90 | 41 |
| ST784 | 5 (7.8) | 1 | 3 | 3 | 2 | 2 | 107 | 3 |
| ST836 | 1 (1.5) | 18 | 48 | 58 | 42 | 4 | 54 | 26 |
| ST852 | 1 (1.5) | 33 | 12 | 40 | 26 | 32 | 157 | 5 |
| ST955 | 1 (1.5) | 21 | 15 | 2 | 28 | 1 | 157 | 4 |
| ST1181 | 1 (1.5) | 33 | 31 | 2 | 28 | 1 | 144 | 5 |
| ST1223 | 1 (1.5) | 10 | 53 | 4 | 64 | 4 | 98 | 5 |
| ST1225 | 3 (4.6) | 1 | 15 | 13 | 60 | 4 | 106 | 2 |
| ST1249 | 1 (1.5) | 1 | 15 | 13 | 60 | 4 | 163 | 2 |
| ST1329 | 1 (1.5) | 1 | 1 | 13 | 12 | 94 | 144 | 2 |
| ST1994 | 1 (1.5) | 56 | 104 | 137 | 98 | 51 | 191 | 74 |
| ST2114 | 2 (3.1) | 1 | 12 | 12 | 64 | 4 | 103 | 3 |
| ND | 1 (1.5) | 1 | 1 | 66 | 60 | 33 | 232 | 41 |
| ND | 1 (1.5) | 32 | 141 | 81 | 43 | 108 | 149 | 45 |
Gene Loci in Acinetobacter baumannii Identified Using Multilocus Sequence Typing
| ST | Antibacterial Susceptibility | |||
|---|---|---|---|---|
| CIP | IMP/MEP (n) | AMK (n) | GEN (n) | |
| ST191 | R | R (23) | R (16) | R (21),I (1) |
| ST229 | R | R | R (1) | R (1) |
| ST369 | R | R | R | R |
| ST451 | R | R | R (2) | R (4) |
| ST784 | R | R | R (4) | R |
| ST1223 | R | R | R | R |
| ST1225 | R | R | S | S |
| ST1249 | R | R | R | R |
| ST1329 | R | S | S | S |
Sequence Typing for Quinolone Resistance in Acinetobacter baumannii
4.6. MALDI-TOF MS Analysis
Several specific peaks ranging from 2,586 - 17,024 m/z were identified in the spectral analysis. A peak was observed at 5,747 m/z for 54 target strains, which is characteristic of A. baumannii. A peak at 9,320 m/z was observed for 52 strains, followed by peaks at 7,434 m/z in 68.8% (n = 44), 4,158 m/z in 62.5% (n = 40), and 12,117 m/z in 48.4% (n = 31) of the strains. Classification based on resistance to major antibiotics using the obtained peak, 78% (n = 50) of the strains with ciprofloxacin resistance showed peaks at 2,586 – 2,827 m/z, 6,091 – 7,273 m/z, 7,464 m/z, 9,092 m/z, and 11,285 m/z. The results were similar to those obtained for imipenem and meropenem, as 78% (n = 50) strains showed resistance to these antibiotics.
5. Discussion
Acinetobacter baumannii is an opportunistic infectious bacterium that causes severe nosocomial infections. The emergence of MDR strains has also been a frequent occurrence in recent years (16). Kumburu et al. (17) reported a result slightly different from this study, with 50% ciprofloxacin resistance in MRAB strain causing infection associated with wounds and pus, which was isolated from hospitals and clinics in Tanzania (Africa) from 2013 – 2015. However, the resistance rates reported for ceftazidime (78.6%, n = 11) and gentamicin (64.3%, n = 9) were similar to those obtained in this study. Sequence analysis was performed to identify potential mutations in the gyrA and parC sequences of the 50 ciprofloxacin-resistant A. baumannii strains.
In gyrA, the codon for serine at the 83rd position was replaced by that for leucine via a point mutation in all the 50 A. baumannii strains. In 86% (n = 43) of the strains, the codon for serine at the 80th position of parC was replaced by the codon for leucine. Lee et al. (18) reported a new point mutation with tryptophan at the Ser-80 position in parC. Thus, high ciprofloxacin resistance was confirmed when the point mutations inducing the replacement of Ser-83 by Leu and Ser-80 by Trp occurred simultaneously. Here, the 43 strains with both gyrA and parC mutations had relatively high MIC values (32 – 256 μg/mL) compared to that of strains with only gyrA mutations.
Meanwhile, MLST analysis of the 64 strains showed the presence of 18 STs, and ST191 accounted for the majority (39%, n = 25) (Table 2). In a study on the gene cassette and molecular typing of A. baumannii integrons isolated from clinical specimens (19), ST195, ST208, ST218, and ST368 were isolated from regions in the vicinity of Shaanxi and Xi’an, China. The STs belonging to CC92 were slightly different from those obtained previously (ST208 and ST1145), suggesting that CC92 exhibited global and regional differences not only in European countries such as Italy, Spain, United Kingdom, Greece, and Denmark, but also in Asian countries such as China, Korea, Thailand, India, and Lebanon. CC92 produces various carbapenemases and is considered the largest and most widely distributed clone worldwide, accounting for the majority of MRAB strains (19). In addition, AUAB08, AUMR02, and AUMR22, identified under ST191, had mutations in both gyrA and parC, and the MICs were as high as 256 μg/mL. In contrast, AUAB22, AUMR17, and AUMR26, also identified under ST191, had mutations only in gyrA, and the MICs were relatively low (32 – 64 μg/mL). This suggests that even within the same ST, differences in sensitivity may occur due to the presence of various ciprofloxacin resistance mechanisms.
Meanwhile, the results of MALDI-TOF MS analysis revealed six common peaks (4158, 4264, 5747, 7434, 8487, and 9320 m/z) in spectra of most of the strains, and characteristic peak of A. baumannii at 5,747 m/z was observed in spectra for all 64 target strains. The major peaks appeared at 9,320, 7,434, and 12,117 m/z, which is in contrast to the previous reports where the main peaks of A. baumannii formed at 4,244, 8,485, and 5,747 m/z (20). Upon classifying the strains (exhibiting different antibiotic resistance patterns) according to their peaks, the spectra of 78% (n = 50) of strains resistant to ciprofloxacin, imipenem, and meropenem were observed to display relatively high intensity peaks at 2,586 – 2,827 m/z, 6,091 – 7,273 m/z, 7,464 m/z, 9,092 m/z, and 11,285 m/z, whereas this pattern was observed in only 19% (n = 12) of the susceptible strains. In a previous study (21), MALDI-TOF MS and antibiotic resistance analysis for A. baumannii showed that the positions of high-intensity peaks vary with the antibiotic resistance patterns. Here, a peak was detected at 2,033 m/z in the spectra of 213 colistin-resistant A. baumannii strain isolated from clinical settings.
5.1. Conclusions
Here, most of the A. baumannii strains exhibited multidrug resistance to various antibiotics, including ciprofloxacin and levofloxacin. Our findings will be useful in antibiotic selection during the treatment of infections caused by MRAB. These findings may also help to determine the effective antibiotic concentration and the methods for preventing group infection and its spread.