1. Background
The Staphylococcus aureus is a Gram-positive bacterium that is normally found as normal flora of the body. It can be found on skin as well as in the upper respiratory system (1). Staphylococcus aureus is an opportunistic bacterium, which can lead to a wide range of infectious illnesses, particularly infections acquired in the community and hospitals. Staphylococcus aureus infection combined with influenza virus infection has been found in recent research to enhance the risk of pneumonia and eventual mortality (2). Methicillin-resistant S. aureus (MRSA)-related antimicrobial resistance necessitates constant efforts to develop plans to eliminate the problem. New treatments and procedures are required to avoid it (3).
Methicillin-resistant S. aureus can cause skin infections, urinary tract infections (UTI), bloodstream infections, food poisoning, and respiratory disorders by producing enterotoxins and alpha pore-forming toxins, which kill host cells and tissues. There are also other factors of S. aureus infection pathogenicity (4). It is commonly seen in many skin disorders and various types, but not limited to a broad range of infections such as recurrent and deep wounds. It has been shown to have the ability to colonize mucosal surfaces (5). If a breach occurs in the mucous membrane or skin, bacteria can move into the tissues or bloodstream (5, 6) S. aureus causes a wide range of diseases, including S. aureus, which may induce bacteremia and endocarditis (7).
Staphylococcus aureus is a polysaccharide-like substance present in many Gram-negative bacteria, a variety of which are frequently involved in biofilm formation. The ability of MRSA to produce biofilm by tissue culture plate (TCP) was investigated, and the results indicated that MRSA isolates showed highly and strong biofilm formation (8). It is synthesized by four different polycarbonate N-acetamide acetic acids (PNAG) into four polymeric polymers (9). The destruction of the icaA genes causes biofilm degradation (8). The gene of icaA is recognized to be encoded the N-acetyl-glucosamine transferase transmembrane synthesizing PNAG polymers (10). Staphylococcus aureus regulates virulence through the network, detects environmental cues, and responds by altering the production of virulence components required for the host to survive.
The accessory gene regulator (agr) is the most prominent quorum sensing QS system among the several S. aureus regulatory systems. It can sense the local concentration of signal molecules, allowing S. aureus to sense its population density and transform this information into gene expression mode (11). Each of these virulence factors activates a quorum-sensing (or titer) device (quantum system), which is referred to as the so-called bacterial sensing peptide (QS) (12).
2. Objectives
The goal of this study is to determine the biofilm-forming ability of clinical isolates from patients in Iraq, as well as the detection of the icaA and agr genes.
3. Methods
This descriptive study describes the ability of S. aureus to resist antibiotics through its production of biofilm.
3.1. Sample Collection
A total of 150 clinical specimens were collected from December 2020 to February 2021 from patients in different hospitals of medical city in Baghdad (Private Nursing Home Hospital and Baghdad Teaching Hospital). Patients' age ranged from 13 to 68 years. Information was additionally taken about the idea of work just as where they do live and whether they experience side effects of different infections. The clinical samples included: (1) wounds, 48 samples; (2) abscess, 35 samples; (3) sputum, 39 samples; and (4) ear infections, 28 samples. The samples were conducted by swab and then cultured on CHROM agar supplied by Condalab Company in Spain, blood agar, and mannitol salt agar manufactured by Oxoid Company (England).
3.2. Isolation and Identification of Staphylococcus aureus
Specifically, the bacteria were tested on a mannitol salt culture, which is unique for S. aureus. Three days after that, the samples were incubated for a further period of time at 37°C. The broth-saline and mannitol salt agar isolates were stored at 4°C before usage. These samples were identified using routine biochemical tests according to according to Abdolmaleki et al. (2019) (13).
3.3. Biofilm Assay
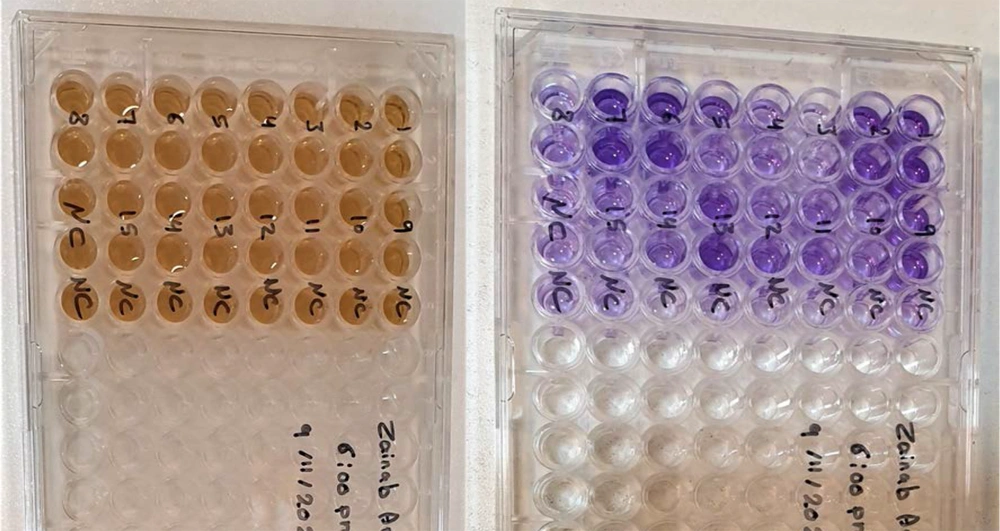
The specificity of S. aureus biofilms was measured. Two wells of a freshly prepared culture brain infusion broth (equivalent to 2x BHI bacteria McFarland No. 5) were added to each sterile 96-well polystyrene microtiter plate. Subsequently, all microplates were incubated at an anaerobic condition (no air) for 24 h. Each sample was replicated three times. Each well was emptied and then washed with 200 μL purified water three times. Next, 200 μL of methane was added, then the dishes were placed in an oven at 60°C for 15 min to remove residual moisture. Crystal violet was applied next, then they were left at room temperature for 5 min. The procedure was followed as described. After finishing the drying, the plates were incubated at 37°C for approximately 30 min afterward. Hence, additional ethanol was introduced at a rate of 200 μL/min for the next 10 min. Finally, each well was calculated at 630 nm with a microplate reader that used visible-light spectrophotometry (14, 15). The results were divided into four categories according to their optical densities as follows: (1) strong biofilm (4 × ODc < OD); (2) middle biofilm producer (2 × ODc < OD ≤ 4 × ODc); (3) weak biofilm producer (ODc < OD ≤ 2 × ODc); (4) non-biofilm producer (OD ≤ ODc).
3.4. DNA Extraction
Genomic DNA was extracted from the clinical isolates of S. aureus 44 isolates utilizing (DNA scaled-down pack that was provided by ABIO organization guidelines, USA) (16).
3.5. Purification and Concentration Measurement
After finishing the extraction process and obtaining pure DNA, the concentration and purity of DNA were measured using a Nanodrop.
3.6. Preparation of PCR Mixture
All DNA extracted from 44 isolates of S. aureus in the current study went through PCR procedure to assess different drug resistance genes. Each PCR reaction had a final volume of 20 μL. Promega master mix was used, and the melted solution was homogenized using a vortex for ten min before using the PCR Master Mix (2x) (Promega company- USA). Homogenizing was done until the priming was done.
3.7. Primer Preparation
Forward and reverse primers, which were in lyophilization status, were dissolved and diluted first in free nuclease double sterile distilled water (amount according to recommended of manufactured company) to obtains 100 pico-mol/μL and this was considered a stock solution, then it was stored in deep freeze. This stock was diluted in free nuclease double distilled water to obtain nearly 10 pico-mol/μL and was stored in deep freeze until used in PCR mixture. It was supplied by Macrogen in a lyophilized suspension. To make a lyophilized 100 pmol/mL solution of the primers, the stock solution was dissolved in nuclease-free water. A working solution of these primers (cooled to -20°C) was generated by combining 10 μL of primer solution with 90 μL of non-nuclease water to yield a final concentration of 10 pMol (Table 1).
3.8. PCR Amplification
The icaA and agr genes were amplified using primers (Table 1). The PCR amplification was conducted in a total volume of 20 μL containing 3 μL DNA, 10 μL Master Mix PCR (Promega Company, USA), 10 pmol of each primer (forward and reverse), and then nuclease-free water was added to a tube to a total volume from 20 μL. Thermo cycling conditions were as follows: (1) initial denaturation at 5 min at 94°C followed by 30 cycles of denaturation 94°C for 30 sec; (2) annealing at 58°C for30 sec; (3) extension at 72°C for 30 sec and a final extension of 72°C for 5 min.
3.9. Agarose Preparation
Agarose (1 g of agar per 100 mL of water previously mixed with 1X TBE buffer and boiled and allowed to cool at 50°C) was applied to the 100 mL solution. A 3 µL of ethidium bromide solution was previously prepared and added to agar. Placing the comb in agarose solution permitted to polymerize (soaked in around 30 min at room temperature), and it was then withdrawn after the agarose solution hardened. The gel was put into the 1X TBE buffer-filled gel tube. It was completely flushed out with buffer (19).
3.10. Sample Preparation
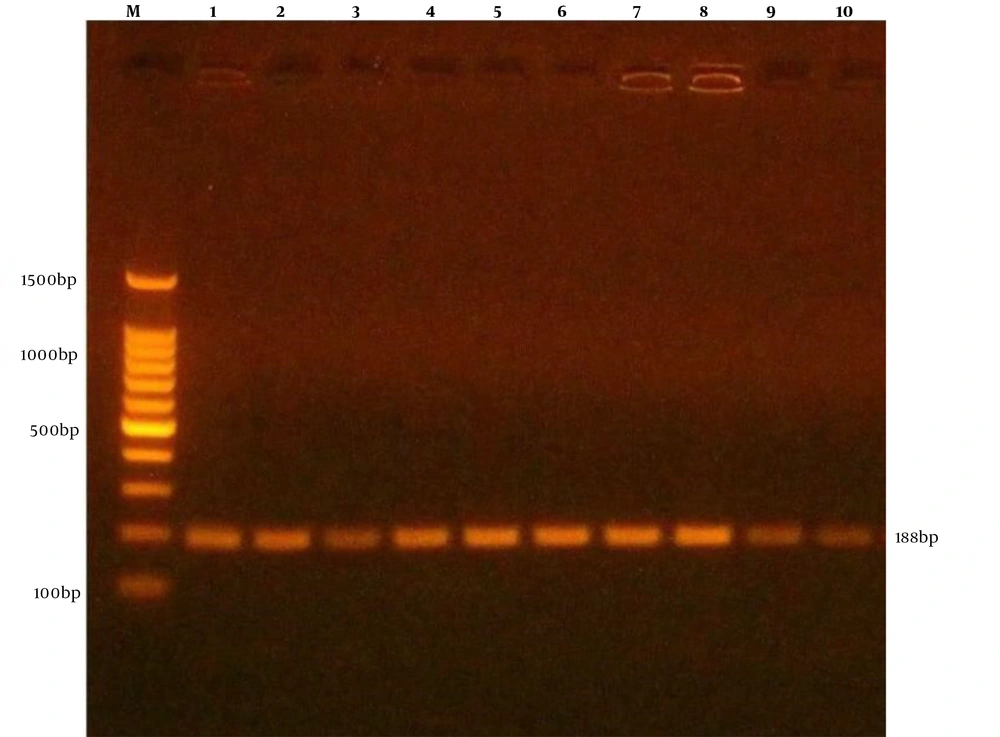
Seven microliters of the PCR product were added to each well of the gel to be used for gel electrophoresis using a DNA ladder (100-bp DNA) (Intron company - Korea). The DNA samples were recorded by ultraviolet transilluminator documentation method (20, 21).
3.11. Statistical Analysis
To recognize the impact of various parts in research parameters, the Statistical Analysis System- SAS (2012) application was used. In this analysis, the chi-square test was utilized to make a significant correlation between rates.
4. Results
4.1. Isolation
The current results showed that of 150 samples, 44 isolates were S. aureus (29.3%), of, wounds samples 45.83% were S. aureus, 37.14% were from abscess, 17.95% from sputum, 7.14% from ear samples (Table 2). All isolates were identified to species morphologically and biochemically based on Bergey’s manual of systematic bacteriology (22). Al-Zoubi et al. (23) showed that most of S. aureus were isolated from wound specimens and abscess. Also, S. aureus has been found to be a common etiology of pneumonia and wounds, resulting from surgical operations that occur often in healthcare facilities, which is the second most prevalent source of bloodstream infections in hospitals (24).
| Sample Sources | No. of Samples | No. of S. aureus Isolates | Percentage |
|---|---|---|---|
| Wounds | 48 | 22 | 45.83 |
| Abscess | 35 | 13 | 37.14 |
| Sputum | 39 | 7 | 17.95 |
| Ear | 28 | 2 | 7.14 |
| Total | 150 | 44 | 29.3 |
| Chi-square (χ2) | - | - | 11.483 a |
a (P ≤ 0.01).
4.2. Biofilm Formation
The results indicated that 47.7% of the isolates produced a strong biofilm, especially in patients between 40 to 68 years, 38.6% produced moderate biofilms, and 13.6% had weak biofilm (Figure 1 and Table 3). This coincides with the study conducted by Muhammad, who addressed biofilm formation by MRSA (25). He presented that 100% of the S. aureus isolates were capable to produce a biofilm. In 2017, Saleh and Khalaf conducted another local investigation. They found that 15% of S. aureus isolates formed weak biofilms, 15% formed moderate biofilms, and 70% formed strong biofilms (26).
| Variables | No. | Percentage |
|---|---|---|
| Weak | 6 | 13.6 |
| Moderate | 17 | 38.6 |
| Strong | 21 | 47.7 |
a Chi-square (χ2) = 9.477 **, (P ≤ 0.01).
4.3. Extraction of DNA
The DNA of S. aureus was extracted using an extraction kit provided by the ABIO Company. In the beginning, all 44 isolates isolated from different sources were grown on mannitol salt agar medium to activate bacteria, which was reserved on the maintenance medium and then transferred to the nutrient broth (Figure 2).
4.4. DNA Concentration and Purity Measurements
The concentration and purity of DNA were assessed by Nanodrop at the end of the DNA extraction process. The authors found a concentration of (75 - 290 ng/L) and purity of (1.65 - 2.2 nm).
4.5. Detection of icaA and agr Genes
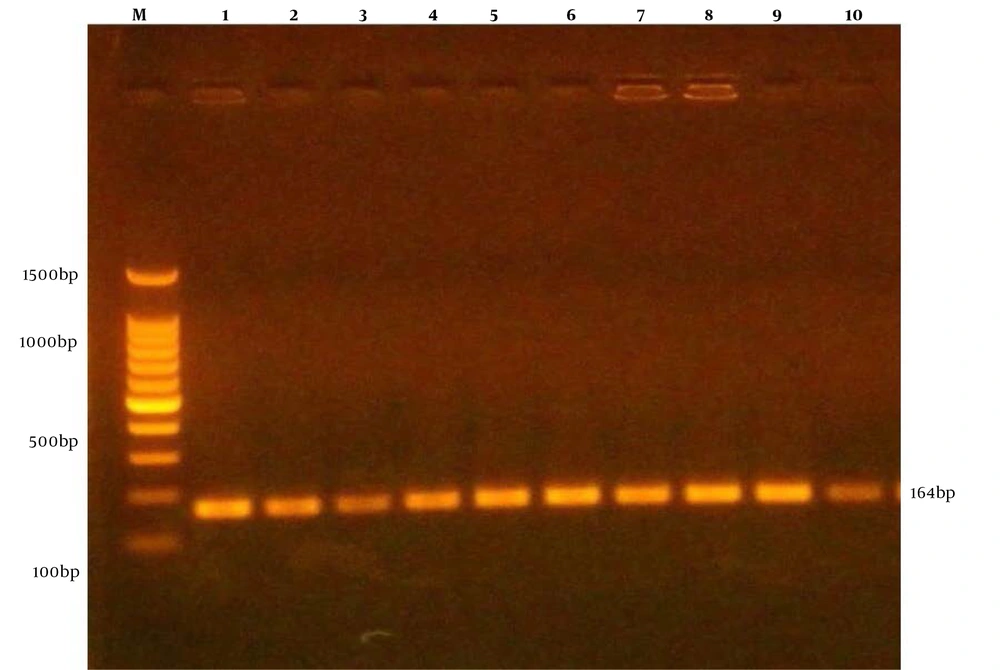
The present molecular results showed that S. aureus from different samples were 13 (59.1%), 4 (30.77%), 3 (42.85%), 0 (0%) from wounds, abscess, sputum, ear, respectively, were positive for agr gene (Table 4 and Figure 3). The results showed that 18 (81.8%), 10 (76.9%), 5 (71.4%), 1 (50%) were from wounds, abscess, sputum, ear, respectively, were positive for icaA gene (Table 4 and Figure 4).
a Values are expressed as No. (%) unless otherwise indicated.
b (P ≤ 0.01).
5. Discussion
Bacterial infection is a critical issue in hospitals and healthcare facilities. Infections caused by MRSA isolates have increased recently, and these infections are more commonly linked to mortality than infections caused by other bacteria (27). Bacterial biofilms are more resistant to antimicrobial treatment and immunological factors from the host (28). In the present study, the agr and icaA genes of S. aureus and their biofilm-forming capacity were studied. The production of biofilm in S. aureus is controlled by icaA biofilm operon and agr locus, the highly dynamic biofilm composition prompting the exchange and removal of nutrient, gas, waste, and solution through channels (29). In an alternative study in 2018, it was confirmed that, in comparison to the planktonic condition for the same MRSA expression, icaA gene expression was significantly increased under biofilm conditions (1). The biofilm-forming processes of S. aureus are determined by the icaA gene cluster that is responsible for the synthesis of polysaccharide intracellular adhesion and capsular polysaccharide adhesion (30). Moreover, S.aureus isolates that were screened from wound infections were positive for icaA gene (31). It was also found that the agr gene was the most frequent biofilm-forming gene among MRSA strains (32).
Biofilm formation was assessed using a tissue culture plate approach in our investigation, and the results showed that 47.7% of the 21 MRSA isolates produced strong biofilms, whereas only 38.6% of isolates formed moderate biofilms. This contradicted the findings of Dakheel et al. (2016) who investigated 25 MRSA strains and found that 88% of isolates produced intermediate biofilms, while 11% produced weak biofilms (33). Another study found that 47.9% of MRSA isolates were moderate biofilm-forming, 39.7% were strong biofilm-forming, and 11% were weak biofilm-forming (34). As a result, it appears that the strains in our investigation are more pathogenic, producing more strong biofilms. This may be attributable to the specimen type or the use of antibiotics in Iraq. Furthermore, the use of inadequately sterilized medical devices resulted in environmental selection in our strains that favored strong biofilm formation. Multiple genes, including SarA and agr genes, regulate the ica genes. They can interact and regulate biofilm creation by interacting with each other. The SarA gene appears to be a master controller of biofilm formation, increasing the synthesis of fibronectin and fibrinogen-binding proteins, as well as toxins for tissue dissemination, while suppressing the expression of protein A (35).
5.1. Conclusions
Most S. aureus isolates that were isolated from wound were biofilm-positive as it was less for the rest of the isolates, and the biofilm formation capacity was weaker. These isolates were bearing icaA and agr genes in a high quantity.