1. Background
Antibiotic resistance is expected to cause more deaths than cancer by 2050, and it poses a severe threat to public health (1, 2). Multidrug-resistant gram-negative bacilli are the most frequent causes of sepsis and pneumonia in newborns, causing death (3, 4). Escherichia coli is one of the most common ones (5-7). There are eight recognized species in the genus Escherichia (8). Escherichia coli is the most studied one (9). Its closest relative is E. fergusonii, with 64% genetic similarity (10, 11). The proximity between the two species gives rise to identification problems, depending on the method used.
With the use of phenotypic methods, E. fergusonii is usually identified as E. coli (12, 13). Epidemiologically, bacterial misclassification leads to underreporting pathogenic microorganisms. Escherichia fergusonii emerges as a microorganism of concern because of its potential for multi-drug resistance; however, it is underreported (13-15). There are three reports of isolation in humans; two are clinical cases (14, 15), and only one reports strains found in different patients (13). Currently, the prevalence and incidence of cases in which the bacterium causes disease are not known precisely. It is also not known exactly how often it becomes resistant to antimicrobial therapy.
2. Objectives
We aimed to present the first report on the isolation and molecular identification of multidrug-resistant E. fergusonii strains (previously identified as E. coli) in infants less than two months old in a neonatal intensive care unit.
3. Methods
3.1. Samples
A descriptive cross-sectional study was performed. Samples of peripheral blood (n = 3), central blood (n = 1), pleural fluid (n = 1), urine (n = 2), and injury discharge (n = 2) were collected from patients under two months of age, admitted to the Neonatal Intensive Care Unit of the Maternal-Perinatal Hospital "Mónica Pretelini Sáenz" of the Instituto de Salud del Estado de México (ISEM) in Toluca, Mexico, from October 2019 to February 2020. Epidemiological data were collected from each patient.
3.2. Culture
All samples were inoculated on the culture media, blood agar (Condalab 1108), Mannitol Salt (BD-BBL 254027), McConkey (BD-BBL 211662), and Biggy Agar (BD-BBL 255002). The specific treatment of each sample was as follows. Blood samples were placed in BacT/ALERT culture medium (Biomerieux 10853) to optimize bacterial recovery. They were subsequently inoculated by a cross streak in the media already described. Pleural fluid and urine samples were centrifuged at 2500 rpm for 10 min, and the sediments obtained were inoculated by cross streak. Injury discharge samples were collected using the "Culture Swab Collection & Transport System" (BBL HFT016) and inoculated by cross streak swabbing. All inoculated culture media were incubated at 35 ± 2°C for 24 h. The macroscopic and microscopic morphological characteristics of each strain were described.
3.3. Phenotypic Identification and Antimicrobial Susceptibility Test with Microdilution Broth Method
Phenotypic identification and antibiotic susceptibility testing with the microdilution method were performed with the MicroScan (autoSCAN-4, Beckman Coulter®) according to the manufacturer's instructions. Neg Combo 67 (Beckman Coulter B1017-421) was used, which included the following antibiotic families: Aminoglycosides, Beta-lactams, Glycyclines, Polymyxins, Quinolones, Sulfonamides, and Tetracyclines.
3.4. Complementary Antibiotic Resistance Test by the Modified Kirby-Bauer Disk Diffusion Method
The strains were subsequently tested for resistance by disk diffusion using the modified Kirby-Bauer method, with the addition of antibiotics not examined in the microdilution test. In addition, antibiotics from the Phenicol, Phosphonate, and Nitrofuran families were added. A pure 18 - 24 h culture equivalent to 0.5 (1.5 × 108 cells) on the McFarland scale was used. Discs with the selected antibiotics were placed in a Müller Hinton agar plate (BIO-RAD 63824) and incubated under aerobic conditions at 35 ± 2°C for 18 - 24 h. For reading, the edge of the halo was visible to the naked eye at the point of complete inhibition from the back of the Petri dish against a black background illuminated with reflected light (16). The diameter was measured in millimeters and interpreted according to the CLSI M100 guide as sensitive (S), intermediate (I), or resistant (R) (16). Multi-resistance was defined as simultaneous resistance to three or more families of antibiotics from a minimum of 10 families (17). The collection strain E. coli ATCC® 25922 was used for quality control (16).
3.5. Molecular Identification
Each strain was seeded in an enriched medium (BHI), and incubated at 35 ± 2°C for 18 - 24 h under aerobic conditions. Biomass was collected, centrifuged at 9,000 rpm for five minutes, and the supernatant was decanted. The Wizard® Genomic DNA Purification Kit (Promega A1120) protocol was used to extract the genomic DNA, and the quality was checked by electrophoresis on a 1% agarose gel under the following conditions: 120 V for 30 minutes with TAE buffer 1X (TAE buffer Invitrogen 5M0311). The concentration was quantified in the EPOCH spectrophotometer (BioTek) at 260 and 280 nm.
Strains were identified by 16S rRNA gene sequencing analysis. Universal primers 27F (5'-AGA GTT TGA TCM TGG CTC AG-3') and 1492R (5'-TAC GGY TAC CTT GTT ACG ACT T-3') were used. The Polymerase Chain Reaction (PCR) was carried out in an Axygen thermal cycler (MaxyGen-II model), using Taq DNA polymerase (Bioline 21105). The conditions were as follows: an initial denaturation cycle of five minutes (94°C), denaturation of one minute (94°C), annealing for 30 seconds (59°C), an extension for one minute (72°C). Besides, 30 cycles were repeated, followed by a final extension cycle for 10 minutes (72°C).
The amplification products were observed in 1% agarose gel electrophoresis under the same conditions used previously, and the concentration was quantified with the EPOCH spectrophotometer. The PCR products were purified using the Wizard® SV Gel and PCR Clean-Up System kit (Promega A9282), and then sent to Macrogen® Sequencing Service (Maryland, USA). Sequences were corrected and assembled using BioEdit v7.2.5 software. They were then compared with BLAST (Basic Local Alignment Search Tool) software and the EzBioCloud public database.
4. Results
The characteristics of the patients and samples are summarized in Table 1. Nine strains showed multi-drug resistance in the microdilution method and disk diffusion antibiograms. Eight of the nine strains were extended-spectrum beta-lactamase (ESBL) producers (Table 2). The two methods showed that all isolates (100%) were resistant to aminoglycosides. Within the beta-lactams, nine strains (100%) were resistant to penicillin and cephalosporins; 77% were resistant to penicillin with inhibitor; strains Uro6 and Uro42 were the only ones resistant to carbapenems (22%). Besides, 100% were resistant to quinolones and sulfonamides, and 88% were resistant to tetracyclines.
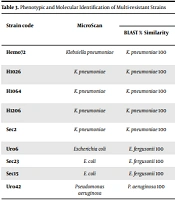
With the microdilution method, sensitivity to tigecycline and colistin was detected in all strains (100%). Ceftazidime/avibactam and ceftolozane/tazobactam combinations showed the best sensitivity results for all strains tested with the disk diffusion method (Table 2). Bacterial identification by phenotypic and molecular methods showed differences in the taxonomic assignment of E. fergusonii. Using molecular identification, strains Uro6, Sec23, and Sec15, phenotypically categorized as E. coli, were identified as E. fergusonii. In addition, five strains were identified as Klebsiella pneumoniae and one as Pseudomonas aeruginosa. These results were consistent with both identification methods. Complete identification data are shown in Table 3.
| Patients | Sex | Age | Diagnosis | Sample Type | Sample Code |
|---|---|---|---|---|---|
| 1 | Male | 60 days | Sepsis | Peripheral blood | Hemo72 |
| 2 | Male | 5 days | Sepsis | Peripheral blood | H1026 |
| 3 | Female | 21 days | Sepsis | Peripheral blood | H1064 |
| 4 | Male | 30 days | Sepsis | Central blood | H1206 |
| 5 | Male | 8 days | Chylothorax | Pleural fluid | Sec2 |
| 6 | Female | 21 days | Sepsis | Urine | Uro6 |
| 7 | Male | 38 days | Transgestational urinary infection | Urine | Uro42 |
| 8 | Male | 14 days | Injury with abscess | Injury discharge | Sec23 |
| 9 | Male | 14 days | Neural tube deficiency | Injury discharge | Sec15 |
Characteristics of the Patients and Samples Included in the Study
| Antibiotic Family | Method | Strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hemo72 | H1026 | H1064 | H1206 | Sec2 | Uro6 | Sec23 | Sec15 | Uro42 | ||
| Aminoglycosides | ||||||||||
| Amikacine | Aa | S | S | S | I | S | S | S | S | R |
| Gentamycin | A | R | S | R | R | R | S | S | S | R |
| Netilmicin | Bb | R | R | R | R | R | R | R | R | R |
| Tobramycin | A | R | S | R | R | R | R | S | S | R |
| Beta-lactam | ||||||||||
| Penicillin | ||||||||||
| Ampicillin | A, B | R | R | R | R | R | R | R | R | ND |
| Piperazine | A | R | R | ND | R | ND | ND | ND | ND | R |
| Penicillin/inhibitor | ||||||||||
| Amoxicillin/clavulanic acid | A | ND | ND | I | ND | I | R | S | S | ND |
| Ampicillin/sulbactam | A, B | R | R | R | R | R | R | S | S | R |
| Piperacillin/tazobactam | A, B | R | S | I | I | S | R | S | S | R |
| Carbapenems | ||||||||||
| Doripenem | B | S | S | S | S | S | R | S | S | I |
| Ertapenem | A | S | S | S | S | S | R | S | S | ND |
| Imipenem | A | S | S | S | S | S | R | S | S | R |
| Meropenem | A | S | S | S | S | S | R | S | S | R |
| First-generation cephalosporin | ||||||||||
| Cefazolin | B | R | R | R | R | R | R | R | R | ND |
| Second-generation cephalosporin | ||||||||||
| Cefaclor | B | R | R | R | R | R | R | R | R | ND |
| Cefotetan | A, B | S | S | S | S | S | R | S | S | ND |
| Cefuroxime | A | R | R | R | R | R | R | R | R | ND |
| Third-generation cephalosporin | ||||||||||
| Cefoperazone | B | R | R | R | R | R | R | R | R | ND |
| Cefotaxime | A, B | R | R | Rc | Rc | Rc | R | Rc | Rc | ND |
| Cefpodoxime | B | R | R | R | R | R | R | R | R | ND |
| Ceftazidime | A | Rc | Rc | ND | Rc | Rc | Rc | Rc | Rc | R |
| Ceftibuten | B | R | R | R | R | R | R | S | R | ND |
| Ceftriaxone | A | Rc | Rc | Rc | Rc | Rc | R | Rc | Rc | ND |
| Third-generation cephalosporin/inhibitor | ||||||||||
| Ceftazidime/avibactam | B | S | S | S | S | S | S | S | S | S |
| Fourth-generation cephalosporins | ||||||||||
| Cefepime | A, B | R | R | R | R | R | R | R | R | R |
| Fifth-generation cephalosporins | ||||||||||
| Ceftaroline | B | R | R | R | R | R | R | R | R | ND |
| Fifth-generation cephalosporins/inhibitor | ||||||||||
| Ceftolozane/tazobactam | B | S | S | S | S | S | S | S | S | S |
| Phenicol | ||||||||||
| Chloramphenicol | B | R | S | R | R | I | I | I | R | ND |
| Phosphonates | ||||||||||
| Phoosphomycin | B | ND | ND | ND | ND | ND | S | ND | ND | R |
| Glycylcicline | ||||||||||
| Tigecyclin | A | S | S | S | S | S | S | S | S | ND |
| Nitrofurans | ||||||||||
| Nitrofurantoin | B | ND | ND | ND | ND | ND | R | ND | ND | ND |
| Polymyxins | ||||||||||
| Colistina | A | S | S | S | S | S | S | S | S | S |
| Quinolones | ||||||||||
| Second-generation quinolones | ||||||||||
| Ciprofloxacin | A, B | R | R | R | R | R | R | R | R | R |
| Third-generation quinolones | ||||||||||
| Levofloxacin | A | S | S | R | S | S | R | R | R | R |
| Sulfonamides | ||||||||||
| Trimethoprim/sulfamethoxazol | A, B | R | R | R | R | R | R | R | R | ND |
| Tetracyclines | ||||||||||
| Tetracycline | A | R | S | R | R | R | R | R | R | ND |
Patterns of Antibiotic Resistance and Sensitivity by Microdilution Method (MicroScan) and Disk Diffusion Method (modified Kirby-Bauer)
| Strain code | MicroScan | Molecular Identification | ||||
|---|---|---|---|---|---|---|
| BLAST % Similarity | Reference Strain | EzBioCloud % Similarity | Reference Strain | Fragment Length | ||
| Hemo72 | Klebsiella pneumoniae | K. pneumoniae 100 | DSM 30104 | K. pneumoniae subsp pneumoniae 99.1 | DSM 30104 | 1500 bp |
| H1026 | K. pneumoniae | K. pneumoniae 100 | DSM 30104 | K. pneumoniae subsp pneumoniae 100 | DSM 30104 | 1438 bp |
| H1064 | K. pneumoniae | K. pneumoniae 100 | DSM 30104 | K. pneumoniae subsp pneumoniae 100 | DSM 30104 | 1456 bp |
| H1206 | K. pneumoniae | K. pneumoniae 100 | DSM 30104 | K. pneumoniae subsp pneumoniae 99.1 | DSM 30104 | 1520 bp |
| Sec2 | K. pneumoniae | K. pneumoniae 100 | DSM 30104 | K. pneumoniae subsp pneumoniae 100 | DSM 30104 | 1425 bp |
| Uro6 | Escherichia coli | E. fergusonii 100 | ATCC 35469 | E. fergusonii 99.92 | ATCC 35469 | 1420 bp |
| Sec23 | E. coli | E. fergusonii 100 | ATCC 35469 | E. fergusonii 99.93 | ATCC 35469 | 1464 bp |
| Sec15 | E. coli | E. fergusonii 100 | ATCC 35469 | E. fergusonii 99.93 | ATCC 35469 | 1462 bp |
| Uro42 | Pseudomonas aeruginosa | P. aeruginosa 100 | DSM 50071 | P. aeruginosa 100 | JCM 5962 | 1452 bp |
Phenotypic and Molecular Identification of Multi-resistant Strains
5. Discussion
Neonatal sepsis is one of the leading causes of mortality in Mexico (3-7). The detection of multidrug-resistant bacteria is essential for infection control, preventing their spread and unfavorable morbidity and mortality implications. The appropriate use of antimicrobials represents positive variables to reduce the burden of neonatal sepsis (18). In this study, E. fergusonii was identified by phenotypic methods as E. coli. However, by molecular techniques, a more precise identification was achieved, coinciding with previous publications that clarified the need to correctly identify this species to complete a taxonomic assignment and accurate epidemiological report (10-13).
Escherichia fergusonii was mainly isolated from injury discharge, except for strain Uro6, which, together with strain Uro42 (P. aeruginosa), was isolated from urine samples and had the highest antibiotic resistance. Both were the only ones resistant to carbapenems. A previous study revealed a similar pattern in septic wound isolates (13). Our results provide new information about the presence of multidrug-resistant E. fergusonii in urine samples.
In 1993, Funke et al. (19) isolated E. fergusonii from human clinical samples and concluded that the bacterium had pathogenic potential; before this, it had not been associated with clinical infections. Current studies on this subject are limited (19-22), and publications on its multi-resistance associated with human pathologies are scarce (13-15). We can affirm that this is the first report of the presence of the bacterium with multidrug-resistant patterns in clinical samples from infants under two months of age admitted to a neonatal intensive care unit.
Strain Uro42, genetically identified as P. aeruginosa, showed resistance to the most significant number of antibiotics tested, including carbapenems. Several studies have addressed the causes of this resistance (23-25). Our results agree with that reported by Mohsen et al. (26), who found 45% carbapenem-resistant Pseudomonas infection in infected neonates. Carbapenems are the last line of defense against many drug-resistant bacterial infections. Unfortunately, our study reaffirms that conditions due to pathogens resistant to this drug are present in this age group. Klebsiella pneumoniae was the most abundant species, isolated from blood and pleural fluid. It is reported as one of the predominant organisms in this type of patient (26-29). The strains were sensitive to carbapenems in contrast to P. aeruginosa and E. fergusonii. Despite being one of the most frequent bacteria in neonatal sepsis, our results showed that its resistance patterns are lower than those found in the other two species reported.
Although the microdilution technique (MicroScan) is most widely used, the disk diffusion methodology (modified Kirby-Bauer) showed important information, such as enzymatic activity (beta-lactamases, metallo-beta-lactamases), resistant mutants, antagonism, and synergism between antibiotics; this provides a broader view in the proper interpretation of the antibiogram. The microdilution method (MicroScan) made it possible to determine that the multidrug-resistant strains were sensitive to tigecycline and colistin. However, these antibiotics are not recommended for children under eight years of age, and colistin is only recommended for children over two years of age (30, 31). With the disk diffusion method, it was possible to determine another sensitivity pattern. Ceftazidime/avibactam and ceftolozane/tazobactam may be more useful in practice, specifically for the age group included in this study.
Respecting the study's limitations, we can mention that it included a small number of cases in a single health care hospital. It is difficult to generalize the results because the pathogens found in neonatal sepsis vary worldwide. The type of sepsis was also not determined; this would have provided important information as some studies showed that late sepsis and early-onset sepsis differ in antibiotic susceptibility patterns, but some studies contradict this claim (29). Among the strengths of our study, we can mention that the latest generation antibiotics were used. Besides, two methods were applied to determine resistance patterns, which allowed us to calculate treatment options according to the age of the patients. Phenotypic and molecular methods were tested for correct bacterial identification, which allowed us to determine the presence of E. fergusonii, rarely reported and with an alarming degree of antimicrobial resistance, in a sensitive age group.
5.1. Conclusions
Phenotypic testing did not allow reliable identification to species level. The 16S rRNA gene sequencing was more accurate for taxonomic assignment. Multidrug-resistant E. fergusonii strains were identified for the first time in children younger than two months. Ceftazidime/avibactam and ceftolozane/tazobactam provided the best treatment option for all strains, considering the age characteristics of the patients. The knowledge generated may promote targeted antibacterial therapy. This study may be of great importance to measure the impact on the local epidemiology of E. fergusonii and antimicrobial and multi-drug resistance.