1. Background
Real-time PCR (RT-PCR) is one of the most useful molecular methods that are currently used in the diagnosis of important infections (1, 2). This method is also used in the recent pandemic of SARS-CoV-2 as the main method for certain diagnoses of this emerging virus (3). Sensitivity and specificity are the main and important advantages of this method, which will be achieved if a precise design and quality material are used (4). Of course, this method might have false-negative results, particularly for the detection of viruses that cause respiratory infections such as SARS-CoV-2, influenza viruses, respiratory syncytial virus (RSV), and other RNA viruses. Obtaining false-negative results depends on various factors: pre-analytical and analytical factors. The pre-analytical vulnerabilities include improper sample collection, sample type selection, sample collection time or inappropriate sample collection, transport and storage of specimens, manual errors, sample contamination, and interfering substances. Analytical issues include inefficient nucleic acid extraction, operator performance, incorrect interpretation of results, PCR machine malfunctioning, insufficient accuracy in design and optimization of qRT-PCR kits such as primers and probe (lack of harmonization), master mix, and controls, especially internal control (4-7).
The most important factors causing false-negative results are as follows: (1) Sample type: Improper collection and preparation of respiratory samples are a set of false-negative factors in performing real-time PCR (8). Specimens such as Bronchoalveolar and sputum specimens are of the best for lower respiratory tract infections, but swab sampling of nasopharyngeal secretions is the most common type of specimen for isolating respiratory viruses compared to samples provided from oropharyngeal secretions, sputum, stool, or blood (9); (2) based on studies related to SARS-CoV-2, the efficacy of virus detection is 89% between days 0 and 4 after the onset of symptoms, and it is reduced by 54% between days 10 and 14 (10).
On average, the detection of the virus is more common in the lower respiratory tract than in the upper respiratory tract (11). Although the Taqman one-step RT-PCR method can detect infected patients in the early stages of the disease, the sampling time is very important in identifying the virus after the onset of symptoms. A sampling at the wrong time will increase the probability of false-negative results in infected patients (10, 12). The viral respiratory samples should be transported via a reliable cold chain along with Viral Transport Media (VTM). It is highly recommended to store samples at 2 to 8°C for 2 to 5 days or -70°C for longer storage (13, 14). As viral RNA is more unstable than DNA, improper transport and storage of samples increase the risk of false-negative results by the Taqman one-step RT-PCR method. To transmit samples containing RNA viruses, the distance between sampling location and test centers should be considered (15).
The quality of viral nucleic acid extraction has a direct effect on real-time PCR results (16). In fact, the amount of extracted genome, methodology, and various extraction protocols will completely affect the Taqman one-step RT-PCR results (17). Due to the emergency caused by the COVID-19 pandemic throughout the world and the lack of mass production monitoring in a short period of time, products related to respiratory sampling, possible variable conditions for storing samples, viral nucleic acid extraction, and virus detection may do not have very good qualities in order to reach diagnostic reliabilities (18, 19). The appropriate diagnostic performance with high accuracy and precision must be considered for preventing routine false-negative results. The performance of many diagnostic kits is not clear due to the lack of gold standards. Therefore, for a definitive diagnosis and preventing false-negative results, negative samples should be repeated with several diagnostic kits. However, this suggestion is not cost-effective. According to some studies, the false-negative rates for SARS-CoV-2 by Taqman one-step RT-PCR test are about 2-39% (20, 21). Hence, we should use commercial kits that have high accuracy for viral genome identification and appropriate standards for design.
It is necessary to hire trained laboratory staff for caring for patients and proper specimen collection to minimize the risk incurred in the sampling procedure and prevent false or inconclusive test results. Moreover, laboratory staff in molecular wards should be trained well and have expertise in molecular technology to meet working demands and prevent vital analytical errors, including incorrect interpretation of results and misunderstanding in instrumental/equipment malfunctioning (22). For designing ultra-specific and effective primers and probes for the target genes of SARS-CoV-2, the conserved sequences of N, E, and RdRP (RNA-dependent RNA polymerase) genes are used in commercial kits (23, 24). As this virus has newly emerged in human societies, the simultaneous detection of two or three genomic regions was applied to detect the virus by the Taqman one-step RT-PCR method. The detection of at least two viral genes and one internal control gene is the main reason for this. In such cases, the concentration of primers and probes is reduced to prevent the formation of primer-dimers which reduce the probability of low-load detection of the target and cause false-negative results (18, 25).
One of the main drawbacks of designing commercial diagnostic kits in the world is endogenous internal control, which has been introduced by the WHO and CDC as an acceptable sequence for use in virus detection kits (26, 27). Endogenous internal control is one of the controllers in Taqman one-step RT-PCR tests that control the correct performance of nucleic acid extraction and cDNA synthesis (28). Normally, in this type of control, a gene whose expression is not affected during the infection will be selected as the target in human samples. In recent years, the human ribonuclease P (RNase P) gene has been used as one of these endogenous internal controls. Subunit p30 of this protein is one type of ribonuclease that causes breakage in RNA and removal of 5’-extra nucleotides from tRNA precursor. This gene is also located on chromosome 10 (29).
Homo sapiens ribonuclease P/MRP subunit p30 (RPP30) has two variants with 4251 bp in length for variant one and 2332 bp in length for variant two, with 14 and 11 exons, respectively (NCBI reference sequence: NM-001104546.2, NM-006413.5). In this article, we examined the proper performance of this approved endogenous internal control and possible defects in the approved CDC primers and probes for RNase P, which can cause false-negative results in suspected respiratory samples. As a result, we designed ultra-specific primers and probes to span an exon-exon junction to avoid the possibility of genomic DNA amplification and detection.
2. Objectives
This study aimed to investigate false-negative results of SARS-CoV-2 one-step RT-PCR tests induced by RNase P RNA as an internal control and ultra-specific primers and probe design strategy to solve the problem.
3. Methods
3.1. Target and Bioinformatics Analysis of Internal Control Primers and Probe
The sequences of primers and probe recommended by CDC to detect the RNase P gene are as follows (30, 31):
RNase P Forward Primer: AGATTT GGACCTGCGAGCG;
RNase P Reverse Primer: GAGCGGCTGTCTCCACAAGT;
RNase P Probe: TTCTGACCTGAAGGCTCTGCGCG.
In order to evaluate the specificity of the CDC primers and probe, we used the NCBI website (National Center for Biotechnology Information) with BLAST software (Basic Local Alignment Search tool) at https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD = Web&PAGE_TYPE = BlastHome.
3.2. Human Respiratory Samples
Thirty respiratory samples were taken from healthy and suspected Iranian patients with influenza A, B, and SARS-CoV-2 viruses who had negative results for the presence of these viruses by the Taqman one-step RT-PCR method. The samples were used to extract viral nucleic acid by employing QIAamp minElute virus spin commercial kit (Qiagen, Germany) following the manufacturer’s instructions. For further confirmation, the quantity of extracted nucleic acid was measured by NanoDropTM 2000 (Thermo ScientificTM, Waltham, Massachusetts, USA). The extracted RNA was stored at -70°C until analysis.
3.3. Ultra-Specific Design of Primer Pairs and Probe for RNase P RNA
Ultra-specific RT-PCR primer pairs and RT-PCR probe design for RNase P RNA (RPR) was performed by Allelel ID 6 software according to the two variants of RPR (NM-001104546.2 and NM-006413.5). Then, designed primers and probes were used to compare with CDC-approved primers and probes (data of this design not provided in this study). After designing the ultra-specific primers and probes, they were synthesized by Metabion Company (Metabion International AG Company, Germany).
3.4. Taqman One-Step RT-PCR
In order to conform to the BLAST results, Taqman one-step RT-PCR with the synthesized primers and probe was performed according to the protocol introduced by CDC and FDA. Real-time PCR was done by using QuantiTect Probe RT-PCR Kit (Qiagen, Germany) as a master mix. The performed schedule on the device (Rotor-gene Q MDx 5plex-Qiagen) is as follows: Reverse transcription at 50°C for 20 min, 95°C for 15 min, denaturation at 95°C for 5 s, and combined annealing/extension at 55°C for 30 s. To examine the binding of the CDC RNase P primers and probe to the genomic DNA, the reaction was performed comparatively without the reverse transcription synthesis step. The final volume of each reaction was 20 µL.
4. Results
4.1. BLAST
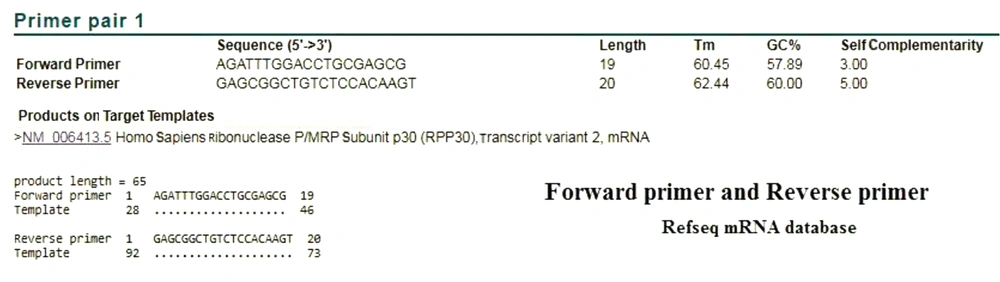
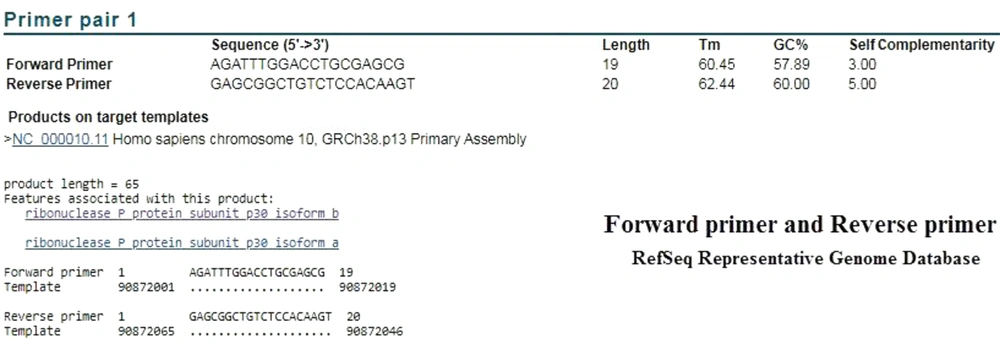
Based on BLAST analysis performed in the Refseq mRNA database, the CDC primers identified both variants of RNase P. The product length was 65 bp, which covered the nucleotides 28 to 92 of mRNA (Figure 1). This area is located in the first exon of the genomic sequence. In order to investigate the specificity of the CDC primers, BLAST analysis was performed in the RefSeq Representative Genome Database to identify possible DNA binding. Surprisingly, the CDC primers of ribonuclease protein subunit p30 were also annealed with chromosome 10, GRCh38.p13, and resulted in a product with the same length and the same annealing parameters (Figure 2).
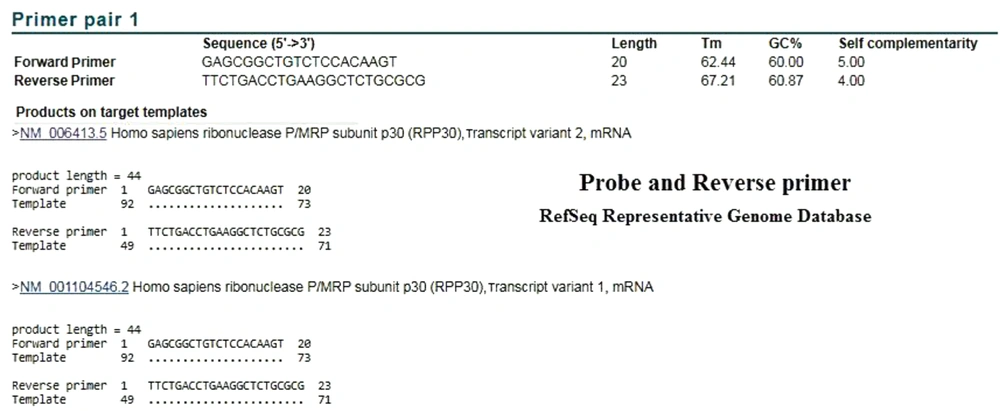
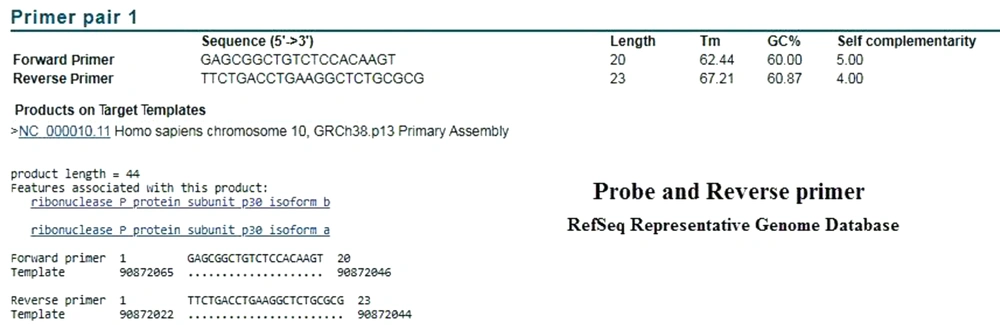
In order to evaluate the performance of the CDC primers and probe, we examined the BLAST analysis in the two mentioned databases. In the analysis of reverse primer and probe in the Refseq mRNA database, both correctly identified the target mRNA. The performed attachment also indicated that the probe was functioning properly, following detection by Taqman one-step RT-PCR (Figures 3 and 4). As expected in either database, no specific attachments were detected in the Forward Primer and Probe mode.
BLAST results on the CDC Reverse Primer and Probe sequences in Refseq mRNA Database. The results showed that the mRNA of RNase P was correctly identified by these two sequences. Unexpectedly, in the RefSeq Representative Genome Database, this attachment was also established on the target DNA (Figure 4).
4.2. Detection of RNase P RNA or DNA/RNA in Taqman One-Step RT-PCR
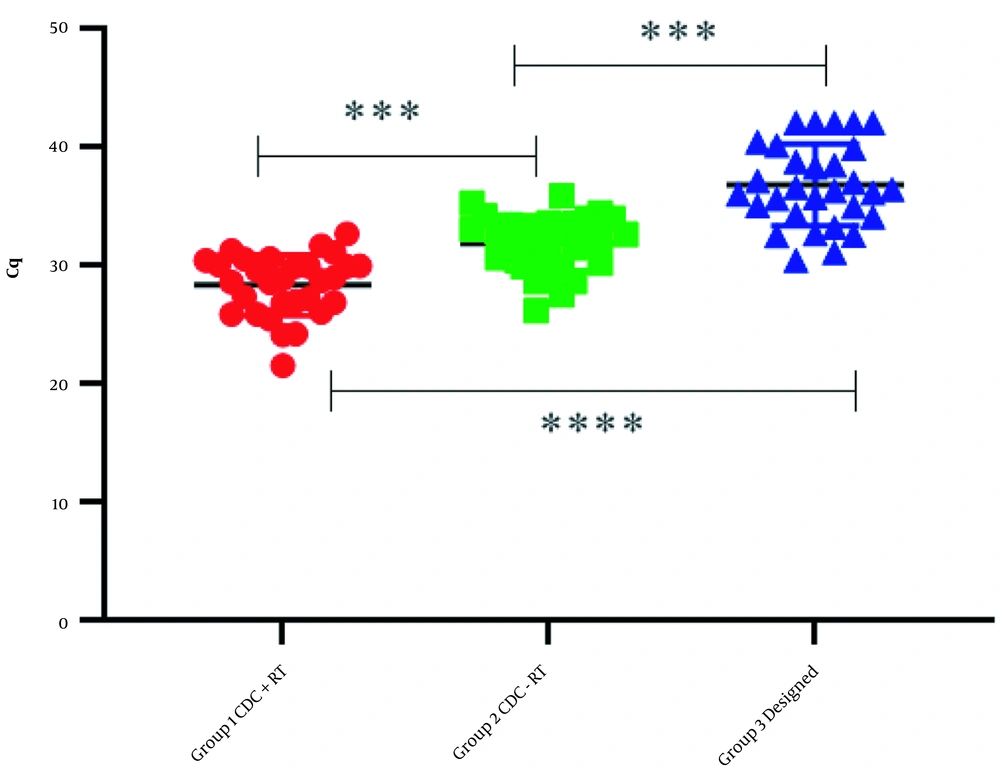
The results obtained from the fluorescent measurement of the RNase P probe indicated that if reverse transcription synthesis was removed from the test steps, the RNase P gene demonstrated a significant Cq with the CDC primers and probe. This indicates binding to the genomic DNA that will invariably cause a false Cq. According to the binding of the primers and probe to the genomic DNA and synthesized cDNA, in the reactions associated with the reverse transcription step, the Cq values of non-specific binding were significant. Therefore, these results prevent the detection of the sufficient RNA present in the initial template. In this study, ultra-specific primers and probes for mRNA RNase P were designed and used for comparative expression. These designed primers and probes did not attach to the genomic DNA of the target gene and did not identify any other non-specific targets (Table 1 and Figure 5).
| Negative Samples for the Presence of Influenza and SARS-CoV-2 Viruses | Group 1: Cq Level in the Program Along with the Reverse Transcription Step with CDC Primers and Probe for RNase P | Group 2: Cq Level in the Program Without Revers Transcription Step with CDC Primers and Probe for RNase P | Group 3: Cq Level in the Program Along with the Reverse Transcription Step with Our Designed Primers and Probe (Ultra-Specific Primers and Probe) | Group 4: Cq Level in the Program Without the Reverse Transcription Step with Our Designed Primers and Probe (Ultra-Specific Primers and Probe) |
|---|---|---|---|---|
| Sample 1 | 25.85 | 28.61 | 32.51 | Negative |
| Sample 2 | 29.90 | 35.22 | Negative | Negative |
| Sample 3 | 26.94 | 29.85 | 38.74 | Negative |
| Sample 4 | 28.67 | 32.60 | Negative | Negative |
| Sample 5 | 29.58 | 33.19 | 40.11 | Negative |
| Sample 6 | 25.87 | 29.42 | 36.39 | Negative |
| Sample 7 | 29.91 | 31.45 | 36.28 | Negative |
| Sample 8 | 32.64 | 34.39 | Negative | Negative |
| Sample 9 | 31.22 | 34.15 | 40.39 | Negative |
| Sample 10 | 30.56 | 33.94 | 39.87 | Negative |
| Sample 11 | 21.54 | 26.15 | 32.64 | Negative |
| Sample 12 | 26.84 | 33.56 | 35.97 | Negative |
| Sample 13 | 31.08 | 33.28 | Negative | Negative |
| Sample 14 | 27.20 | 30.60 | 38.28 | Negative |
| Sample 15 | 29.95 | 32.24 | 36.98 | Negative |
| Sample 16 | 28.84 | 31.47 | 34.05 | Negative |
| Sample 17 | 24.18 | 27.46 | 35.62 | Negative |
| Sample 18 | 28.95 | 33.52 | 35.67 | Negative |
| Sample 19 | 31.64 | 35.84 | 37.12 | Negative |
| Sample 20 | 30.01 | 33.94 | 34.95 | Negative |
| Sample 21 | 28.54 | 31.09 | 30.38 | Negative |
| Sample 22 | 24.12 | 28.62 | 33.20 | Negative |
| Sample 23 | 26.77 | 33.04 | 36.58 | Negative |
| Sample 24 | 30.50 | 32.15 | Negative | Negative |
| Sample 25 | 25.42 | 29.82 | 32.46 | Negative |
| Sample 26 | 27.31 | 31.70 | 34.32 | Negative |
| Sample 27 | 28.55 | 30.94 | 31.07 | Negative |
| Sample 28 | 29.91 | 32.80 | 36.11 | Negative |
| Sample 29 | 30.39 | 33.41 | 38.45 | Negative |
| Sample 30 | 26.02 | 30.15 | 35.00 | Negative |
aCq of the reference is acceptable up to a value of 35.
Based on the results in groups 1 and 2, the RNase P gene detection occurred in the absence and presence of the reverse transcription (RT) step, respectively. This indicates that the signal was false, and CDC primers and probe for RNase P were non-specifically bound to the genomic DNA, which may remain with the routine RNA extraction procedure. According to group 3, the real detection occurred by using the designed ultra-specific primers and probe. Moreover, the expression levels observed in groups 1 and 2 compared to groups 3 were shown to be inappropriate. It is possible that some samples did not have enough RNA; therefore, the test result was reported to be negative for the virus. The absence of non-specific binding of the ultra-specific primers and probe is shown in group 4. In contrast to group 2, where CDC RNase P RNA primers and probe bound to genomic DNA even when the RT phase was omitted, the ultra-specific primers and probe in group 4 did not bind to the DNA even in the absence of the RT phase. Each experiment was performed at least three times.
5. Discussion
Taqman one-step RT-PCR is a definitive method of detecting pathogens, with some advantages and disadvantages (32). One of the main disadvantages of this method is the production of false-negative results. In addition to different test conditions, the correct design of the primers and probe and the quality of master-mix used in the test is very important to prevent false negatives (33). One of the controls used in this method is endogenous internal control, which has a significant controlling role in the correct extraction of nucleic acid and reverse transcription synthesis (34). In this study, we analyzed the endogenous internal control of RNase P, which has been widely used in recent years for influenza and SARS-CoV-2 Taqman one-step RT-PCR tests. Furthermore, it has been approved by reputable organizations for more than a decade.
Based on the results from bioinformatics analysis and Taqman one-step RT-PCR test of CDC primers and probe for mRNA ribonuclease P/MRP subunit p30 (RPP30), this design has been used as an endogenous internal control that attaches to the genomic DNA of RNase P with the same efficacy and the same product size. Therefore, it cannot show the sufficient presence of RNA in the extracted samples and also does not express reverse transcription (cDNA synthesis) correctly. In RNA extraction protocols, the genomic DNA is usually extracted along with RNA, and the existing primers and probe may detect the genomic DNA sequences even in the absence or presence of RNA, which results in non-specific binding.
Generally, internal control is used to check for the presence of appropriate RNA in extracted samples, as well as the reverse transcription step. When there is not enough RNA in extracted samples, but DNA is present, as the CDC primers and probe also bind to DNA, it is wrongly assumed that there is enough RNA in patient samples. Hence, a false attachment of internal control causes a false negative result in the test analysis. However, if the internal control does its work properly and there is not enough RNA in the sample, or even the reverse transcription phase does not work well, the primer and probe will not attach and will not show fluorescence. As a result, a new sample from patients should be taken. Therefore, by a unique primer/probe design from the exon-exon junction, the RNase P gene no longer binds to the genomic DNA non-specifically.
As DNA is more stable than RNA, it becomes more important if samples are not stored properly. Based on the results of the analysis, an obvious error occurred due to the wrong choice of the target sequence region, where the primers and probe are located in the one exon region. In the absence or destruction of RNA or only in the presence of DNA, due to defects in the design of existing primers, the internal control may bind to the genomic DNA non-specifically. Moreover, the level of gene expression is reported to be high in the presence of RNA. According to the protocols of most commercial kits, the RNase P gene must have Cq ≤ 35, which may be due to false amplification when primers and probe bind to the genomic DNA. Therefore, for more accuracy, the sample extraction stage and Taqman one-step RT-PCR testing should be repeated, and it will be more reliable to interpret the results after these repetitions.
Due to the systematic error caused by the type of primers and probe design, laboratories/diagnostic centers are not able to confirm a sufficient amount of RNA in suspected samples. Therefore, negative results cannot be relied on with certainty for the presence of the virus. By using real internal control, re-sampling or RNA re-extraction may be required for some negative results. The CDC suggests that primers and probes can be suspected to obtain false-negative results for the detection of SARS- CoV-2, influenza, and other RNA viruses. Based on the obtained results, the design and use of new primers and probe of internal control are necessary for more accurate tracking of infectious agents, especially respiratory viruses.
5.1. Conclusions
In summary, this study investigated the false-negative results caused by endogenous internal control used in SARS-CoV-2 Taqman one-step RT-PCR detection tests. The evaluation of CDC primers and probe revealed that they are one of the significant reasons for obtaining false-negative results which identify both viral RNA and genomic DNA. However, we designed ultra-specific primers and probe for RNase P RNA (RPR) as an internal control. During SARS-CoV-2 Taqman one-step RT-PCR testing, they do not inappropriately attach to the genomic DNA. Therefore, the non-specific regions of the target gene will not be falsely identified. We highly recommend designing primer pairs and probes across the exon-exon junction in order to achieve high-specific results and minimize false negatives.