1. Background
Bloodstream infection, a serious systemic infectious disease, causes severe sepsis. Each hour of delay in initiation of valid empirical antibiotic therapy was associated with mean increase in mortality of 7.6% (1). In patients with severe sepsis and septic shock, early identification of pathogen and administration of appropriate antibiotic therapy would be conducive to allow the doctors to ensure optimal patient management, which can significantly reduce mortality, hospital costs and prevent the occurrence and development of antibiotic resistance (2, 3). The conventional methods for rapid identification of positive pathogens in blood culture include real time polymerase chain reaction (RT-PCR), matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (MS) combined separation gel SDS, Sepsityper Kit (4-6), etc.; however, PCR method is not sensitive enough to detect pathogens (7), and it depends on some special equipment, manifest a narrow range of diagnostic spectrum (8). The operation of other methods is too complex to be suitable for routine microbial identification.
Blood culture, the gold standard for clinical diagnosis of bloodstream infection, is hindered by slow turn-around time. In the process of traditional methods, time delay from the pathogen incubation and identification to antimicrobial susceptibility testing, requiring at least two days. This impedes early adjustment of valid empirical antibiotic therapy; thus, increasing morbidity and mortality of bloodstream infection patients (9). MALDI-TOF MS has identified pathogens rapidly and accurately by an overnight subculture on solid media. However, there is no reference method for the treatment of samples (10).
2. Objectives
The objectives of the present study were to evaluate the time differences between conventional identification and MALDI-TOF conventional identification and short-incubation MALDI-TOF identification for positive blood cultures and to explore the impact of short-incubation MALDI-TOF identification on empirical antibiotic therapy.
3. Methods
3.1. Study Design
Blood samples were collected from patients with bloodstream infections in laboratory center of Tianjin Medical University General Hospital from 2017 to 2019, only one sample per patient was included in this study. Data were collected from the medical records and laboratory information system. The main information was concluded: the time to initial identification of pathogen, final reporting, antimicrobial susceptibility test, and the initiation of correct empirical antibiotics treatment.
3.2. Instruments and Reagents
3.2.1. Instruments
Matrix-assisted laser desorption ionization time of flight mass spectrometry, MALDI-TOF MS (France BioMérieux), VITEK-2 Compact automatic microbial identification analyzer (France BioMérieux Company), BactecTMFX blood culture instrument and matching blood culture bottle (BD Company, USA), CO2 constant temperature incubator (Thermo Fisher Scientific).
3.2.2. Reagent
VITEK MS-DS target board (France BioMerieux), VITEK MS-CHCA matrix solution (France BioMérieux), Columbia blood agar plate (Zhengzhou Antu Bioengineering Co.Ltd.), MacConkey agar plate (Zhengzhou Antu Bioengineering Co., Ltd.).
3.3. Quality Control Strains
Escherichia coli ATCC25922, E. coli ATCC 8739, Klebsiella pneumoniae ATCC700603, Pseudomonas aeruginosa ATCC27853, Enterococcus faecalis ATCC29212, Staphylococcus aureus ATCC29213.
3.4. Pathogen Identification
In 2017, positive blood culture samples were incubated overnight, then strains were identified by VITEK2 compact automatic bacterial identification system according to colony morphology, Gram staining, and corresponding biochemical reactions. In 2018, pathogens were cultured overnight and identified by MALDI-TOF MS. In 2019, according to the experimental data, the time to incubate of K. pneumoniae, E. coli, P. aeruginosa, Acinetobacter baumannii, and E. faecium were 6 hours, S. aureus and E. faecalis were incubated for 4 hours, pathogen identification was performed by MALDI-TOF MS. MALDI-TOF MS rapid identification process: 1 μL of quality control Escherichia coli ATCC 8739 was added to the steel target, and before bacterial membrane, drying overlaid with 1 μL matrix add, MALDI-TOF MS identification was carried out after natural drying.
3.5. Antimicrobial Susceptibility Test
The antimicrobial susceptibility test was carried out with VITEK2 automated system, and the antimicrobial susceptibility results were interpreted according to the 2020 antimicrobial susceptibility recommendations of the Clinical and Laboratory Standards Institute (CLSI) (11).
3.6. Statistical Methods
Data were analyzed using SPSS (version 23), data whit homogeneity of variance were compared using the analysis of variance, non-parametric test was used for the analysis of variance heterogeneity data. A P-value ≤ 0.05 was considered statistically significant.
4. Results
4.1. Strain Information
A total of 798 strains were collected, 246 in 2017 used conventional identification method, 245 in 2018 after implementation of MALDI-TOF conventional identification, and 289 from 2019 after applied short-incubation MALDI-TOF identification. The strain distribution is shown in Table 1.
| Conventional Identification Method, No. (%) | MALDI-TOF MS Conventional Identification Method, No. (%) | MALDI-TOF MS Short-Incubation Method, No. (%) | |
|---|---|---|---|
| Klebsiella pneumoniae | 76 (30.89) | 72 (29.39) | 78 (26.99) |
| Escherichia coli | 94 (38.21) | 87 (35.51) | 97 (33.56) |
| Pseudomonas aeruginosa | 18 (7.32) | 25 (10.20) | 24 (8.30) |
| Acinetobacter baumannii | 12 (4.88) | 15 (6.12) | 17 (5.88) |
| Staphylococcus aureus | 26 (10.57) | 31 (12.65) | 37 (12.80) |
| Enterococcus faecium | 12 (4.88) | 9 (3.67) | 27 (9.34) |
| E. faecalis | 8 (3.25) | 6 (2.45) | 9 (3.11) |
| Total | 246 (100.00) | 245 (100.00) | 289 (100.00) |
4.2. Results of Three Methods for Identification Time
Compared with conventional identification method, the time to initial identification of K. pneumoniae, E. coli, P. aeruginosa, A. baumannii, S. aureus, E. faecium and E. faecalis decreased by 4.28h, 4.00h, 5.59 h, 5.63 h, 4.63h, 5.92 h and 3.59 h, respectively (P < 0.05) after implementation of MALDI-TOF MS conventional identification and decreased by 22.28 h, 22 h, 23.59 h, 23.63 h, 22.63 h, 23.92 h and 21.59h, respectively (P < 0.05) after applied for short-incubation MALDI-TOF identification, the data are shown in Table 2.
| Conventional Identification Method (h) | MALDI-TOF MS Conventional Culture Method (h) | MALDI-TOF MS Short-Incubation Method (h) | F/χ2 | P | |
|---|---|---|---|---|---|
| Klebsiella pneumoniae | 28.28 ± 0.02 | 24 ± 0.00 | 6 ± 0.00 | 217.43 | < 0.05 |
| Escherichia coli | 28.00 ± 0.38 | 24 ± 0.00 | 6 ± 0.00 | 265.57 | < 0.05 |
| Pseudomonas aeruginosa | 29.59 ± 0.53 | 24 ± 0.00 | 6 ± 0.00 | 64.63 | < 0.05 |
| Acinetobacter baumannii | 29.63 ± 0.52 | 24 ± 0.00 | 6 ± 0.00 | 42.24 | < 0.05 |
| Staphylococcus aureus | 28.63 ± 0.63 | 24 ± 0.00 | 4 ± 0.00 | 90.91 | < 0.05 |
| Enterococcus faecium | 28.92 ± 0.04 | 24 ± 0.00 | 6 ± 0.00 | 46.14 | < 0.05 |
| E. faecalis | 27.59 ± 0.00 | 24 ± 0.00 | 4 ± 0.00 | 36.94 | < 0.05 |
4.3. Results of the Final Report Time
Compared with conventional identification method, the time to final report time of K. pneumoniae, E. coli, P. aeruginosa, A. baumannii, S. aureus, E. faecium and E. faecalis decreased by 29.86 h, 31.20 h, 35.14 h, 34.16 h, 30.30 h, 33.24 h and 28.84 h, respectively (P < 0.05) after implementation of MALDI-TOF MS conventional identification and decreased by 48.85 h, 47.99 h, 55.40 h, 51.07 h, 49.60 h, 51.78 h, 51.73 h, respectively (P < 0.05) after applied for short-incubation MALDI-TOF identification, the data are shown in Table 3.
| Conventional Identification Method (h) | MALDI-TOF MS Conventional Culture Method (h) | MALDI-TOF MS Short-Incubation Method (h) | F/χ2 | P | |
|---|---|---|---|---|---|
| Klebsiella pneumoniae | 64.10 ± 0.17 | 34.24 ± 0.40 | 15.25 ± 0.13 | 199.81 | < 0.05 |
| Escherichia coli | 63.02 ± 2.66 | 31.82 ± 3.42 | 15.03 ± 0.88 | 7165.73 | < 0.05 |
| Pseudomonas aeruginosa | 72.15 ± 3.69 | 37.01 ± 0.24 | 16.75 ± 2.00 | 322.52 | < 0.05 |
| Acinetobacter baumannii | 66.13 ± 1.90 | 31.97 ± 0.51 | 15.06 ± 0.11 | 1618.09 | < 0.05 |
| Staphylococcus aureus | 64.97 ± 1.75 | 34.67 ± 2.34 | 15.37 ± 0.13 | 82.11 | < 0.05 |
| Enterococcus faecium | 67.78 ± 2.18 | 34.54 ± 0.17 | 16.00 ± 0.18 | 144.47 | < 0.05 |
| E. faecalis | 65.89 ± 0.50 | 37.05 ± 0.14 | 14.16 ± 0.13 | 254.20 | < 0.05 |
4.4. Results of Antimicrobial Susceptibility Test Time
Compared with the conventional identification method, the time to antimicrobial susceptibility test of E. coli, A. baumannii, S. aureus in short-incubation MALDI-TOF identification was prolonged by 2.02 h, 2.19 h, 3.86 h, respectively (P < 0.05). Moreover, the time to antimicrobial susceptibility test of K. pneumoniae, E. faecium and E. faecalis was prolonged by 1.71 h, 6.06 h, 3.46 h, respectively; however, this was not significant. In addition, the time to antimicrobial susceptibility test of P. aeruginosa decreased by 1.87 h (P < 0.05). The data are shown in Table 4.
| Conventional Identification Method (h) | MALDI-TOF MS Short-Incubation Method (h) | F/U | P | |
|---|---|---|---|---|
| Klebsiella pneumoniae | 7.54 ± 0.18 | 9.25 ± 0.18 | 1.36 | 0.25 |
| Escherichia coli | 7.01 ± 4.82 | 9.03 ± 1.22 | 44.88 | < 0.05 |
| Pseudomonas aeruginosa | 12.97 ± 3.73 | 11.10 ± 0.16 | 154.00 | < 0.05 |
| Acinetobacter baumannii | 6.87 ± 1.22 | 9.06 ± 1.57 | 31.50 | < 0.05 |
| Staphylococcus aureus | 8.51 ± 0.71 | 12.37 ± 0.19 | 62.72 | < 0.05 |
| Enterococcus faecium | 9.94 ± 3.20 | 16.00 ± 0.18 | 0.00 | 0.94 |
| E. faecalis | 10.70 ± 0.71 | 14.16 ± 0.13 | 0.95 | 0.34 |
4.5. The Coincidence Rate Between the Conventional Identification and Short-Incubation MALDI-TOF Identification Method of Antimicrobial Susceptibility Test
The overall coincidence rate of antimicrobial susceptibility of Gram-negative bacilli was 98.48%, and the general error rate was 1.52%. There were no extremely major errors or major errors, the coincidence rate of antimicrobial susceptibility of all antimicrobials was more than 95%. The overall coincidence rate of antimicrobial susceptibility of Gram-positive bacteria was 99.53%. There was no extremely major error occurred, the rate of major error and minor error was 0.24%, respectively. The data are shown in Tables 5 and 6.
| Antimicrobial | Coincidence Rate, No. (%) | Extremely Major Error, No. (%) | Major Error, No. (%) | Minor Error, No. (%) |
|---|---|---|---|---|
| LevofloxacinK, E | 210 (97.22) | 0 (0.00) | 0 (0.00) | 6 (2.78) |
| CeftazidimeK, E | 215 (99.53) | 0 (0.00) | 0 (0.00) | 1 (0.46) |
| CefepimeK, E | 208 (96.29) | 0 (0.00) | 0 (0.00) | 8 (3.70) |
| ImipenemK, E | 208 (96.29) | 0 (0.00) | 0 (0.00) | 8 (3.70) |
| Cefoperazone SulbactamK, E | 214 (99.07) | 0 (0.00) | 0 (0.00) | 2 (0.93) |
| AmikacinK, E | 212 (98.15) | 0 (0.00) | 0 (0.00) | 4 (1.85) |
| TegacyclineK, E | 216 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Compound sulfamethoxazoleK, E | 216 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| CeftriaxoneK, E | 175 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| CefoxitinK, E | 174 (99.83) | 0 (0.00) | 0 (0.00) | 1 (0.57) |
| ErtapenemK, E | 175 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Amoxicillin clavulanic acidK, E | 168 (96.00) | 0 (0.00) | 0 (0.00) | 7 (4.00) |
| Piperacillin TazobactamK,E | 169 (96.57) | 0 (0.00) | 0 (0.00) | 6 (3.43) |
| AztreonamP, A | 41 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| CiprofloxacinP, A | 41 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| DoxycyclineP, A | 41 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| MinocyclineP, A | 40 (97.56) | 0 (0.00) | 0 (0.00) | 1 (2.44) |
| TobramycinP, A | 41 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| PolymyxinP, A | 41 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| MeropenemP,A | 41 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Total | 2846 (98.48) | 0 (0.00) | 0 (0.00) | 44 (1.52) |
Abbreviations: K, Klebsiella pneumoniae; E, Escherichia coli; P, Pseudomonas aeruginosa; A, Acinetobacter baumannii.
| Antimicrobial | Coincidence Rate, No. (%) | Extremely Major Error, No. (%) | Major Error, No. (%) | Minor Error, No. (%) |
|---|---|---|---|---|
| LevofloxacinS, Efm, Efa | 81 (98.78) | 0 (0.00) | 0 (0.00) | 1 (1.22) |
| PenicillinS, Efm, Efa | 82 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| ErythromycinS, Efm, Efa | 82 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Quinupristine DalfopristinS, Efm, Efa | 82 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| ClindamycinS, Efm, Efa | 82 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| CiprofloxacinS, Efm, Efa | 82 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| MoxifloxacinS,Efm,Efa | 82 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| NitrofurantoinS,Efm,Efa | 81 (98.78) | 0 (0.00) | 0 (0.00) | 1 (1.22) |
| TetracyclineS,Efm,Efa | 82 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| TegacyclineS,Efm,Efa | 82 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| VancomycinS,Efm,Efa | 82 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| LinezolidS,Efm,Efa | 82 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| AmpicillinEfm,Efa | 45 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| GentamicinEfm, Efa | 45 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| StreptomycinEfm,Efa | 44 (98.78) | 0 (0.00) | 1a (0.27) | 0 (0.00) |
| OxacillinS | 36 (97.30) | 0 (0.00) | 1b (5.71) | 0 (0.00) |
| RifampicinS | 37 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| RifampicinS | 36 (97.30) | 0 (0.00) | 0 (0.00) | 1 (2.70) |
| Compound sulfamethoxazoleS | 37 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| total | 1262 (99.53) | 0 (0.00) | 3 (0.24) | 3 (0.24) |
Abbreviations: S, Staphylococcus aureus; Efm, Enterococcus faecium; Efa, E. faecalis
a 1 strain of Enterococcus faecium, the result of short-incubation with high concentration of streptomycin was R, the conventional identification method was S
b1 strain S. aureus, the result of short-incubation method oxacillin was MIC ≥ 4R, and the conventional method was MIC ≥ 0.25s.
4.6. Time to Antibiotic Change
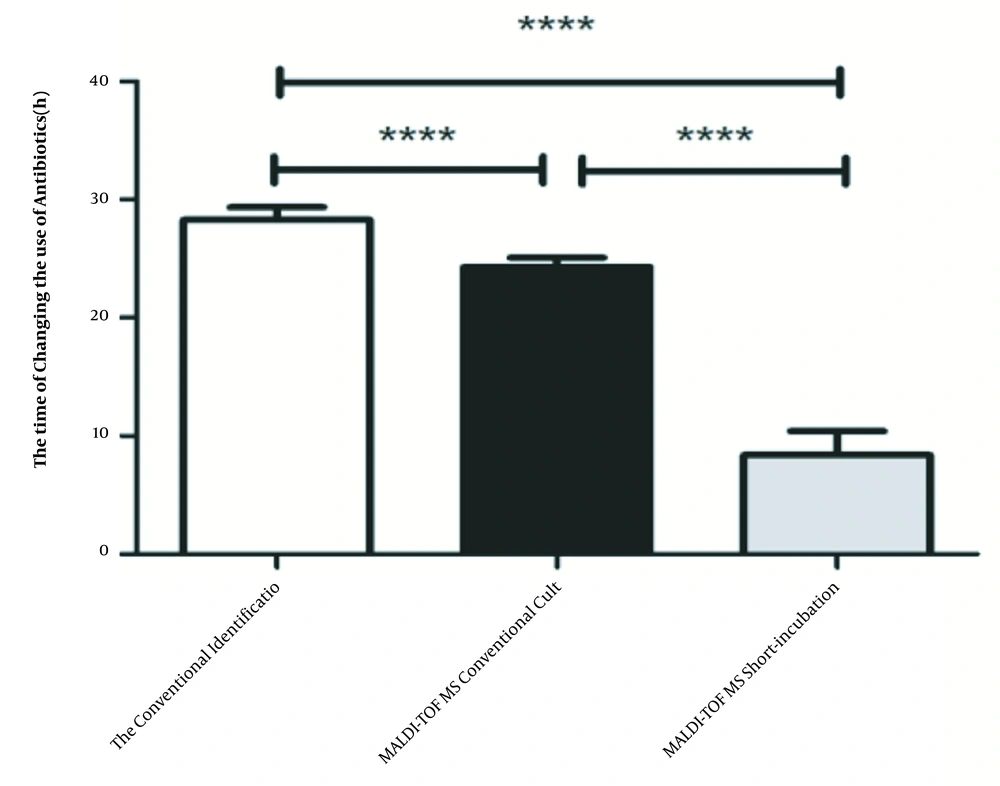
Empirical antibiotic therapy was changed 54.88% (2017), 57.55% (2018) and 62.98% (2019) after pathogen identification. The time to antibiotic change of three methods were 28.27 ± 0.04 h, 24.28 ± 0.35 h, 8.38 ± 1.77 h, respectively. Compared to conventional identification method, the time to antibiotic change was reduced by 3.99 hours (P < 0.001) after the introduction of the conventional MALDI-TOF MS identification, but the statistically significant change occurred after the introduction of short-incubation MALDI-TOF identification (P < 0.001) when patients received more appropriate empirical antibiotic therapy 19.89 hours earlier. The data are shown in Figure 1.
5. Discussion
It could be a critical point in shortening the time of pathogen identification and accurate antibiotic treatment for patients with bloodstream infection as it caused high mortality. Numerous recent studies have already evaluated the capacity of MALDI-TOF MS for rapid pathogen identification when directly used for positive blood culture. Although it has reduced the time to pathogen identification, it is an operating complication for the washing and extraction of bacterial cells and proteins (12), which limits its application in routine microbiological work. Kohlmann et al. (13) showed that MALDI-TOF MS following 4 h short incubation on a solid medium is a valuable tool for rapid pathogen identification from positive blood cultures.
The report (14) demonstrated that MALDI-TOF MS applied to identification of pathogens directly from positive blood culture broths reduced the time to bacterial identification by 28.8 h and increased the proportion of patients on appropriate antimicrobial therapy by 11.3% within 24 h in 253 patients with bloodstream infection. This experiment is based on the main advantage of MALDI-TOF MS to evaluate the time differences between conventional identification and MALDI-TOF conventional identification and short-incubation MALDI-TOF identification for positive blood cultures, and to explore the impact of short-incubation MALDI-TOF identification on empirical antibiotic therapy, in order to improve the level of early diagnosis and treatment of bloodstream infection, reducing the mortality rate.
The results showed that compared with the conventional identification methods, the time to MALDI-TOF MS conventional identification of K. pneumoniae, E. coli, P. aeruginosa, A. baumannii, S. aureus, E. faecium, and E. faecalis decreased by 4.28 h, 4.00 h, 5.59 h, 5.63 h, 4.63 h, 5.92 h and 3.59 h, respectively. The final reporting time decreased by 29.86 h, 31.20 h, 35.14 h, 34.16 h, 30.30 h, 33.24 h and 28.84 h, respectively. The short-incubation MALDI-TOF identification time to initial identification decreased by 22.28 h, 22 h, 23.59 h, 23.63 h, 22.63 h, 23.92 h and 21.59 h, respectively, and the time to final reporting was decreased by 48.85 h, 47.99 h, 55.40 h, 51.07 h, 49.60 h, 51.78 h, and 51.73 h, respectively. Delport et al. (15) found that there was a 16.76 h reduction in time to identification of the pathogen after the introduction of MALDI-TOF identification in 2013, and after implementation of the short incubation MALDI-TOF identification in 2014, the identification time was further shortened by 15.46h. Different from Delport and other studies, in this study, we analyzed the identification time of different strains of common pathogens, determined the identification and final report time of each strain, so as to provide a detailed and accurate report for the early diagnosis and treatment of bloodstream infections.
We found that although short-incubation shortened the identification time, the antimicrobial susceptibility time was not significantly shortened. The time to antimicrobial susceptibility test of E. coli, A. baumannii, and S. aureus in short-incubation MALDI-TOF identification was prolonged by 2.02 h, 2.19 h, 3.86 h respectively, the difference was statistically significant. It is speculated that the reason may be related to the principle of antimicrobial susceptibility test and bacterial maturity of Vitek2 compact automatic bacterial identification system. The principle of Vitek2 compact antimicrobial susceptibility test is mainly to monitor the growth difference of bacteria under antibiotics. The instrument will detect the bacterial growth every 15 min, and the analysis will start only when the bacterial growth reaches the default parameter. If not, the instrument will extend the incubation time.
Due to short-incubation the growth of bacteria may not reach the typical logarithmic growth period (16), the bacterial morphology and physiological activity were not typical, and it is not sensitive to antimicrobial agents, leading to the instrument needs to prolong the incubation time to make the bacterial growth reach the preset parameters, in contrary the growth status and maturity of bacteria cultured overnight are better than those cultured in a short time, and the number of bacteria in the logarithmic growth phase is large, so the antimicrobial sensitivity test can be carried out without prolonging the incubation time. It suggested that although short-incubation shortens the identification time, it relatively prolongs the antimicrobial sensitivity time. Therefore, how to shorten the culture time and antimicrobial sensitivity time is very important for the diagnosis and treatment of bloodstream infection. In this experiment, by comparing the results of short-incubation and routine culture, it was found that the overall coincidence rate of drug sensitivity between Gram-negative bacilli and Gram-positive cocci was more than 98%, and there were no major errors or extremely major errors in Gram-negative bacteria. One strain of E. faecium in Gram-positive cocci made a major error to a high concentration of streptomycin and one strain of S. aureus to oxacillin. The reason may be related to the growth state of bacteria and the principle of instrument detection, and the specific reasons need to be further studied.
Antibiotic treatment was changed 54.88% (2017), 57.55% (2018), and 62.98% (2019) after pathogen identification in three methods. The time to empirical antibiotic change was reduced by 19.89 hours after the introduction of short-incubation MALDI-TOF MS, making doctors significantly advance the time of accurate antibiotic treatment of patients. Kock et al. (17) assessed the effect of short-incubation MALDI-TOF MS identification on empirical antibiotic treatment, found that nearly 20% of antibiotic treatment was adjusted after bacterial identification, and 72% of the adjustment revealed an improvement in the patient's illness condition. Short-incubation MALDI-TOF MS identification is relative to shorten the length of hospital stay and mortality risk, especially in bloodstream infections caused by P. aeruginosa, Enterococcus spp., and AmpC-producing Enterobacteriaceae, short-incubation MALDI-TOF MS identification resulted in 12.8% increase in cases receiving appropriate empirical antibiotic treatment within 48 h (18). Another study (19) established that inappropriate empirical antibiotic treatment is associated with increased mortality and length of hospital stay. So, early identification of specific pathogens in suspicious sepsis or bacteremia and initiation of appropriate antibiotic therapy may significantly improve the outcome of patients with sepsis (2, 20).
5.1. Conclusions
The identification of MALDI-TOF short incubation significantly shortens the time of identification of the pathogen and the time of final exposure, is a reliable method for the rapid identification of positive blood cultures which lead significantly to the earlier appropriate empirical treatment of bacteremia.