1. Background
Herpes simplex virus (HSV) infection is a global health concern. Primary HSV infections mostly occur during infancy, and thereafter the virus remains in the latent form in sensory local ganglia throughout life (1). Primary and recurrent infections with HSV-1 may result in significant psychological and physical morbidities. Herpes simplex virus can be reactivated periodically and usually causes a mild, self-limiting infection in immunocompetent individuals (2). Conversely, patients who are immunosuppressed and immunocompromised are often at high risk of chronic diseases associated with severe HSV-1 infection (1, 3, 4). Acyclovir (ACV) is a synthetic purine analog and the drug of choice for systemic treatment and prophylaxis of HSV-1 infections. The active form is ACV triphosphate. Acyclovir is phosphorylated by both cellular kinases and viral thymidine kinase (TK) and thereafter acts as a DNA chain terminator during replication (5-7).
Acyclovir-resistant isolates affect a significant proportion of immunosuppressed patients but a few (< 1%) immunocompetent individuals with HSV-1 infections (8, 9). This discrepancy could be explained by the presence of functional immune responses in the latter individuals and the diminished pathogenicity of ACV-resistant viruses. Recent studies have suggested that reduced ACV susceptibility of HSV-1 arises in approximately 2.5% to 10.9% of patients who are immunocompromised. The highest prevalence of ACV resistance (25%) was observed in patients who underwent hematopoietic stem cell transplantation (HSCT) (10).
Herpes simplex virus-1 TK is responsible for converting ACV to its active form, and mutations in this gene can lead to ACV resistance. Resistance is associated with mutations in the TK gene in 95% of the cases (5-7, 9, 11). In 5% of the cases, resistance arises from mutations in DNA polymerase (pol), an enzyme responsible for the elongation of viral DNA, leading to altered enzyme activity. Mutations can result in dysfunctional protein structures or a change in substrate specificity (6, 7, 11). Resistance-associated mutations in both TK and pol, including substitution, deletion, or insertion of nucleotides, are particularly prevalent in the active site regions (7, 9, 11).
Very few studies have assessed the ACV susceptibility of clinical isolates from Iranian patients with HSV-1 infection. The HSV-1 resistance to antiviral agents like ACV is primarily evaluated via the characterization of viral phenotype using plaque reduction assays (12, 13). Genotypic tests are used to track resistant viruses based on the presence of resistance-associated mutations in TK and pol (7). One challenge in such analyses is that a considerable number of nucleotide substitutions can be detected, which must be characterized to determine the mutations causing resistance. The interpretation of mutations related to frameshift is easy because it leads to the synthesis of short or non-functional viral enzymes. By contrast, single nonsynonymous substitutions are more difficult to interpret given the high degree of polymorphism in viral genes, particularly TK (7, 12). Thus, genotypic data for HSV-1 isolates with resistant phenotypes are crucial to determine treatment course.
2. Objectives
The present study aimed to characterize 26 HSV-1 clinical isolates from outpatients and organ transplant recipients in terms of phenotypic ACV resistance. Moreover, genotypic resistance to ACV was assessed through sequencing the viral TK and pol genes amplified from virus-infected cell DNA.
3. Methods
3.1. Patients and Viral Strains
Herpes simplex virus-1 clinical isolates were collected from patients who were immunocompetent and immunocompromised with recurrent HSV infections between 2016 and 2019. Patients were referred to the Prof. Alborzi Clinical Microbiology Research Center at Namazi Hospital in Shiraz, Iran. Immunocompromised patients such as solid organ transplant with recurrent HSV infections and immunocompetent individuals in different age groups were included in this study. HIV patients and pregnant women were excluded from the study. Herpes simplex virus-1 samples were tested to verify the susceptibility to ACV using virology laboratory methods.
The diagnosis of HSV-1 infection was developed by polymerization chain reaction (PCR) as described previously (14). Herpes simplex virus-1 isolates were initially propagated on Vero cells (African green monkey kidney) obtained from the Pasteur Institute of Iran (Tehran, Iran). Vero cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Grand Island, NY, USA) supplemented with 1% penicillin/streptomycin (Gibco) and 10% fetal bovine serum (Gibco), at 37°C in 5% CO2. When the titer reached between 106 and 108 Plaque Forming Units (PFU) after a maximum of five passages as explained below, viral vials were prepared and stored at -70°C until analysis. Besides, A549 cells (obtained from the Pasteur Institute of Iran) were used to assess ACV susceptibility. Culture conditions for A549 cells were the same as those for Vero cells. Also, ACV was purchased from Sigma-Aldrich (St. Louis, MO, USA).
3.2. Viral Titer Determination Using Plaque Assay
A standard plaque assay was used to determine the viral titer. Viral culture methods were described previously (15). Vero cells were grown in six-well plates and allowed to reach 70% confluency. The cells were inoculated with serial dilutions of HSV-1 (101 - 106) and incubated for one hour at 37°C under a humidified atmosphere containing 5% CO2. The cells were overlaid with a medium containing agar (1.01615.0100 Merck, Germany), incubated for 48 hours at 37°C under a humidified atmosphere containing 5% CO2, and then fixed with 12.5% formaldehyde. Plaques were examined visually and quantified by crystal violet staining. Plaque assays were carried out in duplicate.
3.3. Differentiation of HSV-1 and HSV-2
A multiplexed real-time PCR for HSV genotyping for HSV-1 and HSV-2 was performed in a single reaction as described previously with modifications (14). Briefly, the optimized gB primer pairs (GbTypF: 5’-CGC ATC AAG ACC ACC TCC TC-3’, GbTypR: 5’- GCT CGC ACC ACG CGA-3’) amplified both HSV-1 and HSV-2 with equal efficiency. The two probes were labeled with different fluorescent dyes. The HSV-1 typing probe (GbTyp1:5’- TGG CAA CGC GGC CCA AC-3’) was fluorescein amidite (FAM) and at the 3’-end with Black Hole Quencher VR (BHQ)-1 (Merck, Darmstadt, Germany). The HSV-2 typing probe (GbTyp2: 5’-CGG CGA TGC GCC CCA G-3’) was labeled at the 5’-end with Yakima Yellow (Generi Biotech, Hradec Kra’lov, Czech Republic) and at the 3’-end with BHQ 1. Reactions were carried out using a StepOnePlus Real-Time PCR instrument (Applied Biosystems, Foster City, CA, USA). Amplification of HSV DNA was performed in a 20.0 µL reaction using qPCRBIO Probe Mix Hi-ROX (PCR Biosystems, London, UK). Reactions contained 10 µL of 2X qPCRBIO Probe Mix, 0.8 µL of primers, 0.8 µL probe, and 5 µL of the extracted DNA sample. After two minutes of polymerase activation at 95°C, the PCR mixture was subjected to 45 cycles of 95°C for 5 s and 60°C for 20 s.
3.4. Phenotypic Analysis
Herpes simplex virus-1 isolates were analyzed phenotypically using the reduction of cytopathic effect (CPE) as the read-out. Briefly, A549 cells (5 × 105 cells per well in 100 mL of DMEM) were placed into a white 96-well plate (SPL Life Sciences, Pocheon, South Korea) and incubated at 37°C under a humidified atmosphere containing 5% CO2 for 24 hours. Based on previous studies (16, 17), three concentrations of ACV (0.01, 0.05, and 0.1 µg/mL in assay media) were selected and added to the plates (one concentration per well). Before infection, each well was incubated with ACV at 37°C for one hour to allow the cells to adapt to the new environment. Then, 10 µL of diluted virus (multiplicity of infection 0.1) was added to each well. In control wells, 100 µL of assay medium was added. The final assay volume was 100 µL per well. Plates were incubated at 37°C for 48 hours. Afterward, the plates were stored at -70°C.
Three freeze-thaw cycles were used to obtain HSV-1. For titer reduction assays, Vero cells (2 × 105 cells in 100 mL of DMEM) were seeded in each well of a white 96-well plate (SPL Life Sciences) and incubated at 37°C under a humidified atmosphere containing 5% CO2 for 24 hours. Serial dilutions of each well for the treatment of A549 cells were prepared in 96-well plates separately. From each serial dilution, 25 µL was pipetted into one row of a 96-well plate, and 75 µL of assay medium was added so that the final assay volume was 100 µL/well. The plates were incubated at 37°C for 48 hours. The cells were fixed with 300 µL per well of 10% formaldehyde and stained with 0.5% crystal violet in 20% methanol. A susceptibility response study (with the virus) was performed to determine the concentration of ACV that inhibited HSV-1-induced CPE. Herpes simplex virus-1 was titrated, and a standard plaque assay was performed on Vero cells using three concentrations of ACV as described above (16). The percentage of inhibition (%I) was calculated for each concentration as follows: %I = (1 - number of viral plaques in treated wells/number of viral plaques in virus control) ×100.
3.5. Genotypic Analysis
The viral DNA was obtained from 200 µl of viral vials using the Viral Nucleic Acid Extraction kit (Roche Diagnostics, Basel, Switzerland). The TK gene, 1417 bp, was amplified in two fragments with sizes of 701 and 716 bp. The pol gene, 3,708 bp, was divided into five fragments for amplification. Each fragment covered about 1,000 bp in the pol gene. The primer sequences were described previously (18). The fragment 701 (TK-R2) primer was modified to amplify the sequence of HSV-1 reference strain 17 (GenBank accession no. X14112) (Table 1).
The PCR reaction mix included 10 µM of each primer, 10 µL of Taq DNA Polymerase 2x Master Mix RED - Ampliqon, and 5 µL of extracted DNA in a volume of 25 µL. Following a first denaturing step for three minutes at 95°C, the PCR mixture was performed (35 cycles) as follows: denaturation at 94°C for 60 s, annealing at 55°C for 60 s, and final elongation at 72°C for 90 s. The reactions were subjected to a final extension step at 72°C for 10 minutes. Amplicons were directly sequenced by Codon Genetic group (Tehran, Iran) using an Applied Biosystems 3130XL instrument. Sequence alignment and comparison with a previously published HSV-1 reference sequence (GenBank accession no. X14112) were performed using MEGA X (www.megasoft ware.net). All nucleic acid sequences were aligned with nucleotide positions in this reference strain. Genotypic analysis of HSV-1 resistance was done through amplified fragments of the viral pol and TK genes, as well as further sequencing.
| HSV DNA Fragment (Amplicon in bp) | Function | Sequence 5 | Nucleotides of HSV Genome |
|---|---|---|---|
| HSV-1 TK | TK-1 | TTTTATTCTGTCCTTTTATTGCCGTCA | 46,607 - 46,634 |
| Fragment 1 (701) | TK-R2 | CATCGCCGCCCTCCTGTGCTACCC* | 47,308 - 47,285 |
| HSV-1 TK | TK-2 | ACGATGTTTGTGCCGGGCAAGGTC | 47,192 - 47,215 |
| Fragment 2 (716) | TK-R1 | CGAGCGACCCTGCAGCGACCCGCT | 47,907 - 47,884 |
| HSV-1 DNA pol | HSV-1-pol-1 | ATCCGCCAGACAAACAAGGCCCTT | 62,655 - 62,678 |
| Fragment 1 (1041) | HSV-1-pol-R4 | CCCCACCCTCGTACTTCTTGATGG | 63,695 - 63,672 |
| HSV-1 DNA pol | HSV-1-pol-2 | GTCCGAAGCGGGCGTGTGCTGTCG | 63,623 - 63,646 |
| Fragment 2 (1033) | HSV-1-pol-R2 | GGCCGTCGTAGATGGTGCGGGTG | 64,655 - 64,633 |
| HSV-1 DNA pol | HSV-1-pol-3 | CCATCTGGAGCTCTCGGCCGTCGC | 64,588 - 64,611 |
| Fragment 3 (1157) | HSV-1-pol-R3 | GCTAAAACAGCAGGTCGACCAGGG | 65,744 - 65,721 |
| HSV-1 DNA pol | HSV-1-pol-4 | AGATGCTCATCAAGGGCGTGGATC | 65,652 - 65,675 |
| Fragments 4 and 5 (1043/1046) | HSV-1-pol-4a | GTAAGATGCTCATCAAGGGCGTGGATC | 65,649 - 65,675 |
| HSV-1-pol-R1 | GGCTCATAGACCGGATGCTCAC | 66,694 - 66,673 |
a*, Modified primer.
3.6. Phylogenetic Analysis of TK and Pol Sequences
Sequences were edited using BioEdit version 7.2 (https://bioedit.sofware.informer. com). Prior to phylogenetic analysis, gaps in the Clustal W genomic alignment were removed leaving 126,608 bp in the alignment. Neighbor-joining phylogenetic trees were produced in MEGA software version X using the Maximum Composite Likelihood model with 50 bootstrap replicates. To understand the evolutionary relationships, the sequences of 26 strains isolated from patients with suspected HSV infection in Fars Province were evaluated. A consensus tree was prepared with a 70% confidence threshold based on the 50 bootstrap replicates for each of the 26 HSV-1 isolates.
4. Results
4.1. Phenotypic Characterization of Acyclovir Susceptibility
Twenty-six HSV-1 clinical isolates were obtained from 11 women and 15 men. The patients’ age ranged between 13 days and 78 years (mean ± standard deviation, 32.1 ± 21.2 years; median, 26 years). Samples were collected using swabs from different anatomical sites. Clinicians provided the following clinical diagnoses: herpes facialis (n = 17), herpes labialis (n = 4), and gingivostomatitis (n = 5). There was no information regarding antiviral therapies received by any of the 26 patients. All 26 HSV-1 isolates induced CPE in Vero cells. All HSV clinical isolates were identified as HSV-1 by PCR genotyping. Susceptibility to ACV was first evaluated on A549 cells, an adherent epithelial human cell line, and CPE was observed. Inhibition of virus infectivity of Vero cells in the presence of ACV was assessed by comparing the viral titer in ACV-treated wells and untreated control wells. All 26 HSV-1 isolates were nearly 100% susceptible at ≥ 0.01 µg.ml-1 ACV. Seven and two isolates, respectively, showed 70% and 80% susceptibility at ≥ 0.05 µg.ml-1 ACV. Two isolates from patients who received transplants and one isolate from an outpatient showed 70% susceptibility at ≥ 0.1 µg.ml-1 ACV.
4.2. Genotypic characterization of Susceptibility Mediated by TK and Pol Gene Mutations
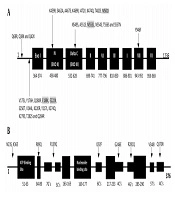
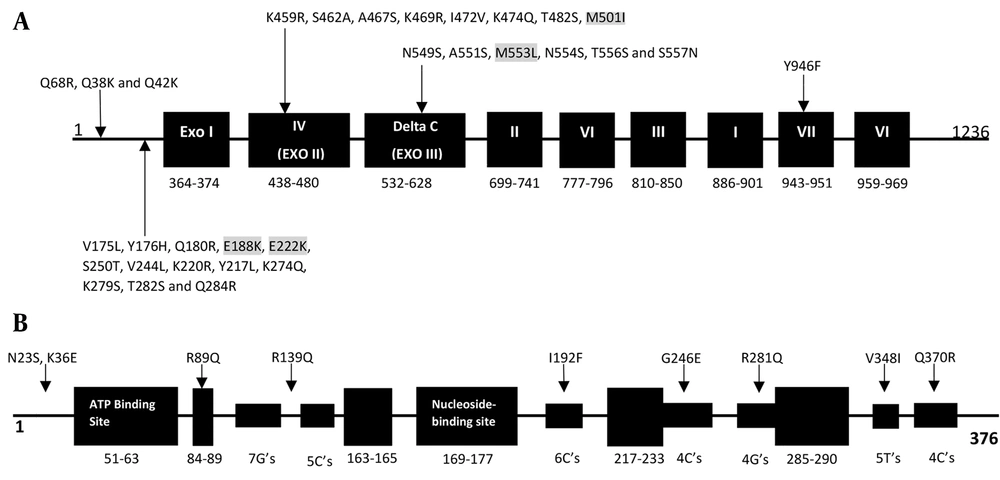
Genotypic results were obtained using amplified fragments and sequencing of the TK and pol genes. The results are shown in Tables 2 and 3. In pol, 31 amino acid substitutions, compared with the reference strain without a premature stop codon, were detected. These substitutions were the result of natural gene polymorphism. The 31 amino acid substitutions were detected in six isolates from patients who received transplants and were associated with the conserved region IV (K459R, S462A, A467S, K469R, I472V, K474Q, T482S, and M501I), the C region (N549S, A551S, M553L, N554S, T556S, and S557N), the VII region (Y946F), and noncoding regions (V175L, Y176H, Q180R, E188K, E222K, S250T, V244L, K220R, Y217L, K274Q, K279S, T282S, Q284R, Q68R, Q38K, and Q42K). Four of the 31 amino acid changes (M553L, M501I, E188K, and E222K) were previously associated with ACV susceptibility (Table 2 and Figure 1A). Surprisingly, the remaining 20 isolates from immunocompetent individuals showed no substitutions in comparison with the HSV-1 reference strain 17. In the TK gene, all 26 HSV-1 isolates contained polymorphism-associated non-synonymous mutations. The N23S and R281Q mutations were observed in all isolates. The K36E mutation was observed in one isolate, V348I was observed in the ninth isolate, R89Q was observed in the sixth isolate, Q370R was observed in the 11th isolate, G246E was observed in the fifth isolate, and I192F and R539Q were both observed in the fourth isolate (Table 3 and Figure 1B). Frameshift and nonsynonymous mutation did not confer susceptibility to ACV.
| Number | Isolation Sites | Polymorphism-Associated Mutations | Susceptibility-Associated Mutations |
|---|---|---|---|
| 2 | Herpes facial | Q68R, Q38K, Y946F | - |
| 3 | Herpes facial | Q42K, V175L, Y176H, Q180R, S250T, V244L, K220R, Y217L, Q284R | - |
| 5 | Herpes facial | K459R, S462A, A467S, K469R, I472V, M501I | M501I |
| 6 | Herpes labialis | N549S, A551S, N554S, T556S, S557N, M553L | M553L |
| 11 | Herpes facial | K274Q, K279S, T282S, E188K | E188K |
| 21 | Herpes facial | K474Q, T482S, E222K | E222K |
| Number | Isolation Sites | Polymorphism-Associated Mutations | Susceptibility-Associated Mutations |
|---|---|---|---|
| 1 | Herpes facial | K36E, N23S, R281Q | - |
| 4 | Herpes facial | I192F, R539Q, N23S, R281Q | - |
| 5 | Herpes facial | G246E, N23S, R281Q | - |
| 6 | Herpes facial | R89Q, N23S, R281Q | - |
| 9 | herpes labialis | V348I, N23S, R281Q | - |
| 11 | Herpes facial | Q370R, N23S, R281Q | - |
4.3. Phylogenetic Analysis
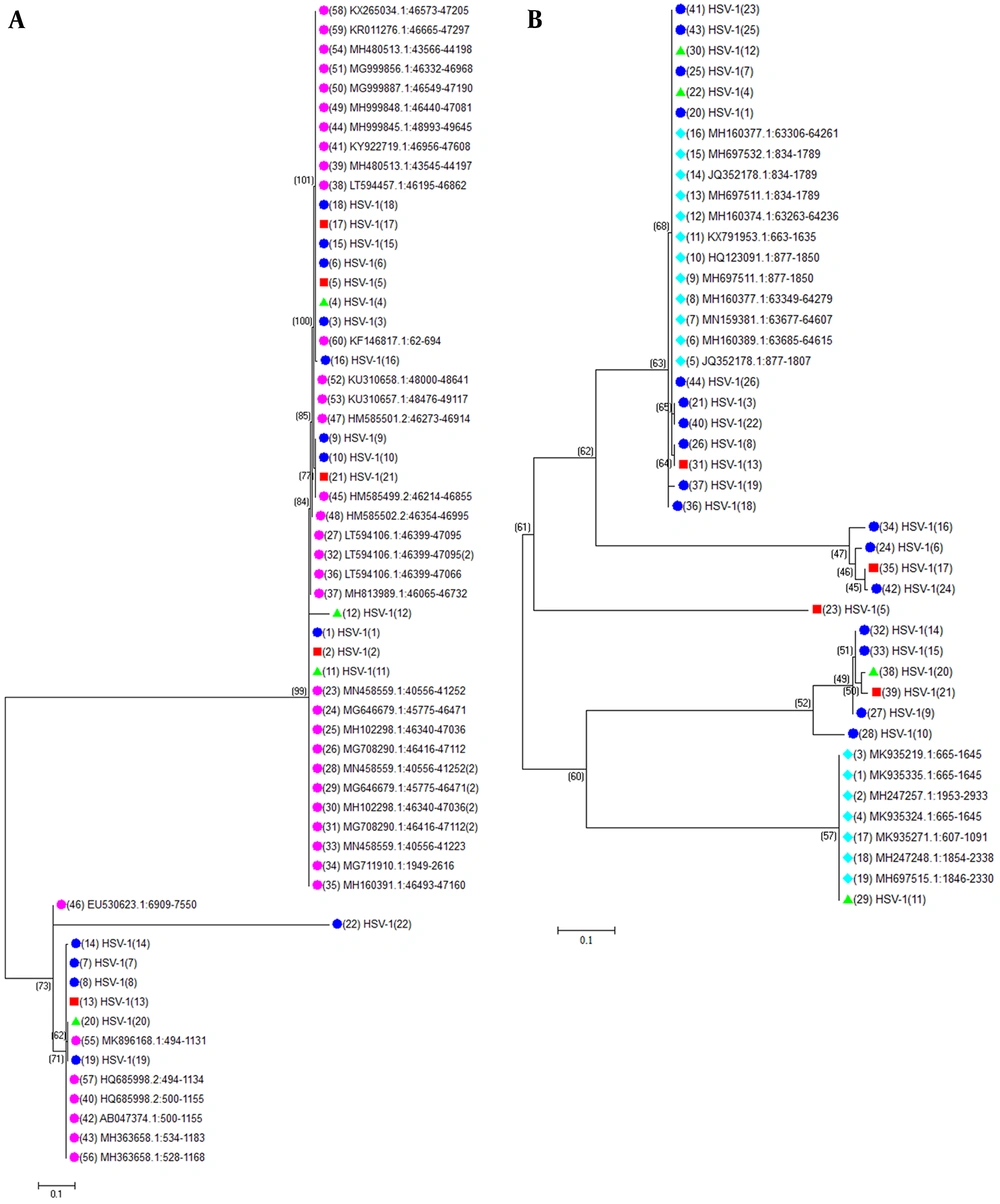
To better understand the molecular evolution of the HSV-1 isolates, the full-length pol and TK genes from the 26 HSV-1 isolates in this study were sequenced. The sequences were evaluated with recorded sequences of HSV-1 TK and pol in the NCBI database. The pol and TK sequences of the HSV-1 isolates showed high nucleotide identity. The TK and pol genes had nucleotide identities of 97.0% to 100% and 96.2% to 100%, respectively. Phylogenetic analysis of these genes revealed that all isolates belonged to the HSV-1 species (Figure 2A and B). The evolutionary relationship of the TK and pol genes suggested harbored greater genetic diversity compared with reference sequences.
Evolutionary relationships of the 26 HSV-1 partial genomic sequences of thymidine kinase (TK) (A) and DNA polymerase (pol) (B). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood (ML) method and are in the units of the number of base substitutions per site. The ML tree was generated from aligned TK, and DNA pol gene sequences using Mega X. Viral isolates are highlighted in different colors based on isolation sites as follows: herpes labialis: light blue; gingivostomatitis: red; and herpes facialis: green
5. Discussion
For decades, nucleoside analog ACV has been the gold standard antiviral therapy for HSV infections (19). However, the ACV-resistant HSV is on the rise with the increasing number of patients who are immunocompromised (20). Mutations are detected in the HSV TK and pol HSV that can give rise to resistance. However, before a clinical isolate is considered resistant, it must be characterized using phenotypic and genotypic characterization assays. The virus present in clinical specimens must first be passaged in tissue culture as the virus can replicate in many cell types (13).
In this study, 26 HSV-1 isolates with possible susceptibility to ACV were analyzed by genotypic and phenotypic methods. According to our study, resistance to ACV is conventionally assessed by phenotypic assays to evaluate the antiviral drug concentration inhibiting 50% of viral replication in vitro. Only three of the 26 HSV-1 isolates examined in our study were susceptible to ACV ≥ 0.1 µg/mL. One isolate was from an outpatient, and the other two were from patients who received transplants. We also determined the TK and pol genes of the 26 HSV-1 isolates. We found that several substitutions in the TK (N23S, R281Q, K36E, V348I, R89Q, Q370R, G246E, I192F, and R139Q) did not contribute to ACV resistance. The N23S and K36E substitutions outside the conserved or active site regions have never been associated with resistance.
The substitutions V348I and Q370R were also not located in the hot spot of the TK gene, and no change in TK function was observed. Most observed substitutions in the TK gene were previously observed in other studies (7, 8, 16, 18, 21, 22). Kudo et al. (18) reported a substitution like Q89R. The following amino acid substitutions in TK were previously identified in HSV-1 clinical isolates: T350S, R89H, R293W, G61A, and I78F. Three reports in the literature have documented 15 HSV-1 isolates containing the substitutions R41H and assessed their ACV sensitivity. Frobert et al. (8) showed that nucleotide substitutions placed in the TK noncoding regions may be associated with ACV resistance, but their role needs to be determined in future experiments. The TK gene is mutated in approximately 95% of ACV-resistant isolates (7, 21).
Our results revealed 31 amino acid replacements related to natural polymorphism and 12 amino acid substitutions unaccompanied by a premature stop codon in pol. Unexpectedly, only one resistance-related substitution (S557N) in the conserved Exo III region of pol was categorized as a natural polymorphism. Sauerbrei et al. (23) reached the same result (i.e., all HSV-1 and HSV-2 isolates had at least one non-synonymous substitution associated with polymorphism in the pol gene). In the current study, we identified two amino acid substitutions situated outside of the conserved region. One amino acid substitution was identified in the conserved region of Exo III (M553L). Our phenotypic study demonstrated that substitutions in this region do not affect viral DNA replication. Gibbs et al. reported that amino acid substitutions in the Exo III region have a significant impact on pol function, leading to impaired replication (24). Our results do not support these findings. Surprisingly, the amino acid substitution (M553L) located in the Exo III region conferred ACV susceptibility in the phenotypic study; all the three substitutions located outside of the conserved regions of pol possessed a sensitive phenotype against ACV.
Data from our study and others suggest that when comparing genotypic and phenotypic results, unreliable correlations can be established between substitutions and their impacts on ACV susceptibility. Previous studies showed that natural polymorphisms in the pol and TK genes occurred similarly in 1973 and 2014 (16, 23, 25). However, a few small changes could be detected in the amino acid sequences of sub-regions of these proteins. Our results indicate that the HSV-1 pol gene has a considerable degree of polymorphism. Because of its large size, HSV-1 pol shows greater genetic variability in most studies because it has a larger number of intragenic polymorphisms. Based on these findings, we conclude that the frequency of ACV-resistant HSV-1 infection has increased over the last few decades, especially in patients who receive transplants. However, our findings indicate that ACV susceptibility occurs predominantly in immunosuppressed patients. In four of 20 patients who were immunocompromised, ACV-susceptible HSV-1 isolates were identified in the genotypic analysis.
In addition, the phylogenetic analysis of partial genome sequences revealed that they belonged to the TK and pol genes of HSV-1. In 2014, Frobert et al. (26) reported a rise in the prevalence of HSV strains resistant to ACV (from 14.3% to 46.5%) among patients with HSCT comparing 2002 to 2006 and 2007 to 2011 periods (8, 26). In other studies, 12 (4.3%) HSV strains resistant to ACV were reported from 276 patients who were immunocompromised; the overall prevalence of ACV resistance was low (between 0.3% and 0.7%) (10, 27-29). A previous study (30) reported that HSV isolates resistant to ACV were identified in up to 4.0% among children who were immunocompetent with oral herpetic lesions. Also, Duan et al. (22) identified ACV-resistant strains of HSV-1 in 6.4% of patients with recurrence of herpes keratitis. According to the literature, the prevalence of ACV-resistant HSV among patients who are immunocompromised is close to 5%, and may reach 14% among HSCT recipients. The results of the present study showed that the number of mutations in HSV-1 TK and pol has increased in patients who receive transplants and undergo long-term treatment compared with outpatients.
The clinical definition of resistance of HSV isolates based solely on genotyping results is not straightforward. Obtaining sequential HSV isolates from a single patient may overcome some of the diagnostic difficulties of ascribing novel point mutations to natural polymorphism or resistance. Our study had several limitations. First and foremost, we did not collect information on patients who were immunocompromised and immunocompetent and their antiviral therapy circumstances. Our primary goal was to conduct the genotyping of pol and TK in 26 HSV-1 isolates over four years. Unfortunately, no solid conclusions can be drawn regarding the emergence and development of ACV resistance within this period because of the limited number of ACV-susceptible strains detected.
5.1. Conclusions
We analyzed 26 HSV-1 isolates obtained within four years and identified natural amino acid polymorphisms in TK and several ACV susceptibility-associated mutations in pol. The considerable HSV-1 genetic variation, which was higher in pol than in TK, makes the rapid and easy genotypic diagnosis of HSV-1 susceptibility. Nevertheless, ACV-susceptible strains were detected predominantly in immunocompromised patients, particularly those undergoing solid-organ transplantation. Mutation accumulation may lead to the appearance of ACV-resistant HSV strains in such patients. The prevalence of ACV-resistant HSV-1 strains in such patients has increased over the past two decades.