1. Background
Salmonella is one of the main foodborne pathogens in the Enterobacteriaceae family. Salmonella spp. cause diarrhea, vomiting, abdominal colic, pneumonia, and bacteremia (1). On the other hand, in the cutting-edge technology of bacterial therapy, scientists utilize bacteria such as Salmonella typhimurium and Shigella flexneri as antiproliferative and potential chemotherapeutic agents to treat gastrointestinal cancers (2, 3). Livestock and poultry products are susceptible to Salmonella contamination. Foods leading to disease outbreaks are mainly eggs and poultry meat; however, the predominant foodborne diseases are enteritis and typhoid fever (4, 5). Due to its excellent survival characteristics, Salmonella is found in various fresh vegetables and meat products. When adhered to the surface of heat-treated foods, the bacteria rapidly are multiplies even at low temperatures (6). Accordingly, the detection of Salmonella is of utmost importance in the food industry (7).
Routine culture is the gold standard for detecting, isolating, and identifying Salmonella spp. However, the cultural process is time-consuming. Samples should be placed in an enrichment solution for 18 - 24 h, transferred to chromogenic plates for 18 - 24 h, and purified for 18 - 24 h. Following these steps, the pathogen can be preliminarily and serologically identified before its identity is confirmed. In this regard, it takes 6 - 7 days to reach results, and specialized laboratories are required (8). Compared with routine culture, polymerase chain reaction (9), DNA microarrays (10), and antibody assays (11) are more appropriate detection methods; however, they are time-consuming. Accordingly, we need a more efficient and straightforward protocol.
In 2000, loop-mediated isothermal amplification (LAMP) was proposed as a novel approach to molecular identification (12). LAMP is specific, sensitive, and cost-effective. However, it fails to distinguish between live and dead cells, and this method often obtains false-positive results from food samples (13). Accordingly, a more accurate method is required to detect and monitor pathogens. Studies have reported that reverse transcription LAMP (RT-LAMP) may overcome the LAMP limitations. RT-LAMP can be used to detect viruses (14). In this study, we collected samples from Hangzhou, Zhejiang Province, China, and used RT-LAMP to detect Salmonella to facilitate the rapid detection of pathogenic bacteria in food.
2. Objectives
To design six primers and detect Salmonella using RT-LAMP to facilitate the rapid detection of the pathogen in food.
3. Methods
3.1. Strains and Culture
We collected 41 strains from different areas in Hangzhou, China. Ten Salmonella spp. were used as positive samples; other bacterial species were selected to evaluate the specificity of RT-LAMP (Table 1). Salmonella strains were stored in the LB broth/agar medium, and additional species such as Listeria were stored in the BHI broth/agar medium. The bacterial solutions were cultured at 37°C and diluted. We measured bacterial concentrations by ultraviolet spectrophotometry. All chemicals and media were acquired from Sangon Co., Ltd. (Shanghai, China).
| Types and Species | Strains |
|---|---|
| Target- Salmonella spp. | |
| Salmonella enterica | ZFM New1 |
| S. enterica | CGMCC 1.1552 |
| S. enterica | CGMCC 1.10603 |
| S. typhimurium | CGMCC 1.1174 |
| S. typhimurium | CGMCC 1.1190 |
| S. pullorum | ZFM 124694 |
| S. anatum | ZFM New2 |
| Non- Salmonella spp. | |
| Micrococcus luteus | CGMCC 1.193 |
| Staphylococcus aureus | ATCC 6538P |
| S. aureus | CGMCC 1.2386 |
| S. aureus | CGMCC 1.879 |
| S. aureus | CGMCC 1.128 |
| Lactobacillus plantarum | CGMCC 1.11 |
| L. plantarum | CGMCC 1.124 |
| L. plantarum | CGMCC 1.551 |
| L. plantarum | CGMCC 1.556 |
| L. plantarum | CGMCC 1.511 |
| L. lactis | ATCC 15577 |
| Bacillus subtilis | CGMCC 1.1627 |
| Enterococcus faecalis | ZFM BM13 |
| E. faecalis | CGMCC 1.125 |
| Shigella flexneri | CGMCC 1.1868 |
| Pseudomonas aeruginosa | CGMCC 1.647 |
| S. dysenteriae | ATCC 9753 |
| Escherichia coli | CGMCC 1.1580 |
| E. coli | O104 |
| E. coli | JM109 |
| Listeria seeligeri | ZFM 51335 |
| L. monocytogenes | ATCC 7648 |
| P. putida | CGMCC 1.645 |
| Rhodotorula Rubra | CGMCC 2.1034 |
| Saccharomyces cerevisiae | CGMCC 2.1643 |
Bacterial Strains Used in this Study
3.2. RNA Extraction and cDNA Synthesis
We extracted total RNA from all 41 bacterial strains using an RNA Rapid Extraction Kit (Sangon Co., Ltd. Shanghai, China) and measured yield using a NanoDrop spectrophotometer (Eppendorf, Germany). To minimize the RNA degradation, total RNA was converted into cDNA using a PrimeScript II First Strand cDNA Synthesis Kit (TaKaRa, China) (15). The reaction volume (20 μL) consisted of 6 µL of RNA, 4 μL of 5× RT buffer, 2 μL of dNTPs (10 mM each), 50 pmol of random hexamers, 10 U of AMV reverse transcriptase XL (TaKaRa), and 10 U of RNase inhibitor (TaKaRa). The reaction solution was maintained for 60 min at 42°C and 15 min at 70°C.
3.3. Primer Design
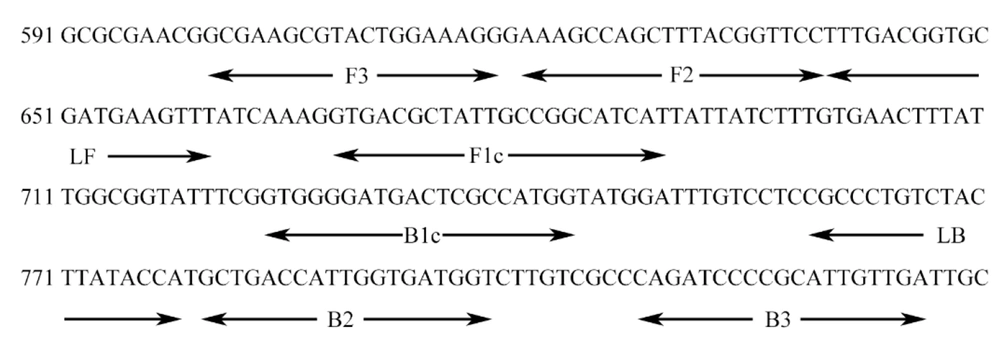
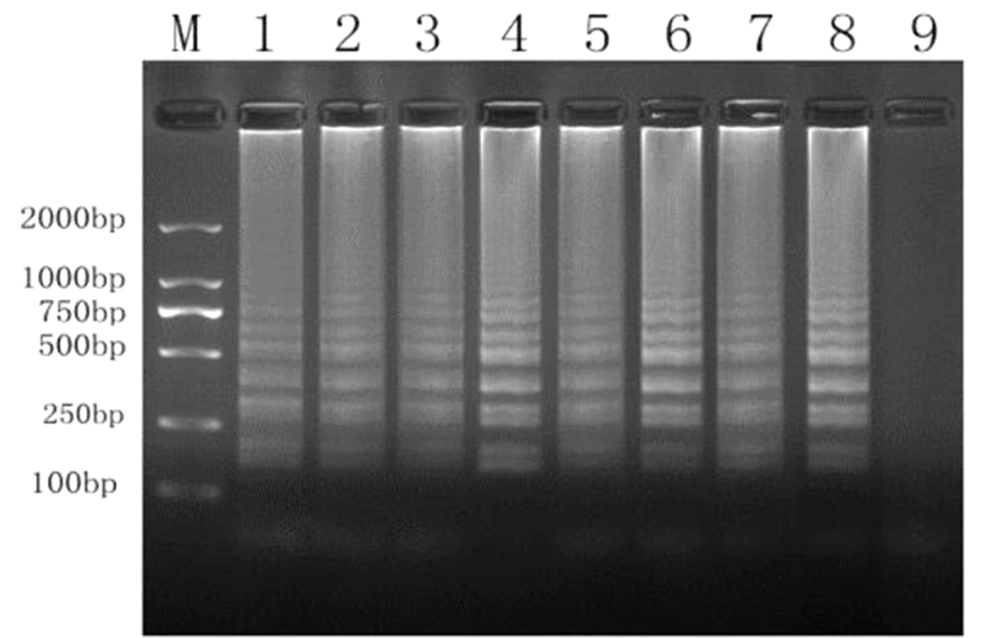
We designed primers based on an internally transcribed spacer sequence (GenBank Accession No. NC_987035) using Primer Explorer V5 (Eiken Chemical Co. Ltd., Tokyo, Japan). Six pairs of primers were created, which targeted distinct regions of the LAMP sequence and corresponded to two outlying primers (F3 and B3), two internal primers (FIP and BIP), and two loop primers. Table 2 and Figure 1 show the primer sequences and locations, respectively.
| Primer Name | Type | Sequences (5' - 3') | Length (bp) |
|---|---|---|---|
| F3 | Forward outer | GCGAAGCGTACTGGAAAGG | 19 |
| B3 | Reverse outer | TCAACAATGCGGGGATCTG | 19 |
| FIP | Composed of F1c and F2 | ATGATGCCGGCAATAGCGTCACAAAGCCAGCTTTACGGTTCC | 40 |
| BIP | Composed of B1c and B2 | GTGGGATGACYCGCCATGGACCATCACCAATGGTCAGC | 38 |
| LF | AAACTTCATCGCACCGTCAAA | 21 | |
| LB | CCGCYCTGTCTACTTATACCA | 21 |
Primers Positions of inv A Gene Coding Sequences of Salmonella
3.4. LAMP Assay
The following six primer pairs were added to a 25-μL tube: 1.6 mM dNTPs, 0.8 M betaine (Sigma-Aldrich, USA), 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 4 mM MgSO4, 0.1% Triton X-100, 80 U Bst DNA polymerase (New England Biolabs, USA), and 2 μL of target cDNA. Different assay conditions, including different temperatures (59 to 65°C) and concentrations of MgSO4 (2 to 8 mM), dNTPs (0.8 to 2.0 mM), and Bst DNA polymerase (40 to 320 U), were evaluated. The reaction solution was incubated for 1 h and 5 min at 80°C (16, 17). All experiments were performed in triplicate.
3.5. Detection of RT-LAMP Products
The amplified products were separated using 2% agarose gel electrophoresis and analyzed under ultraviolet light after 40 min. A white precipitate (magnesium pyrophosphate) was produced during the reaction, which was visible with the naked eye.
3.6. Specificity of RT-LAMP
We used the RNA templates of the 41 strains to perform RT-LAMP. A total of 25 non-Salmonella strains were tested to assess potential cross-reactions (Table 1). The other strains were used for determining the specificity of the assay.
3.7. Sensitivity of RT-LAMP
We determined the detection limit of RT-LAMP by preparing a 10-fold dilution series (106 CFU/μL to 100 CFU/μL) of Salmonella (ZFM New1) in sterile phosphate-buffered saline. The strain was measured by plating it onto LB agar directly (Huankai, Guangzhou, China) followed by incubation at 37°C for 24 h. The RNA templates were obtained from 100 μL of each dilution, and 2 μL fractions were amplified using RT-LAMP. Each sensitivity test was performed four times.
3.8. Salmonella Detection in Ready-to-Eat Meats
We used the RT-LAMP assay to detect Salmonella spp. in ready-to-eat (RTE) meats and other foods and compared the results with those obtained from the Chinese National Standard assay (GB 4789.30-2016). The RTE meats were obtained from a local market in China. We divided each sample into 25-g portions, and each portion was isolated for the RNA extraction to act as a template for RT-LAMP. The GB assay was performed by incubating 25 g of each sample at 30°C for 24 h in 225 mL of Listeria enrichment broth I (LB1). In the next phase, we incubated 100 μL of the samples in 10-mL of Listeria enrichment broth II (LB2) at 30°C for 24 h. Positive LB2 cultures aliquots (50 μL) were inoculated on PALCAM agar (Huankai) and Listeria chromogenic agar (Huankai) and incubated at 37°C for 48 h. To confirm the presence of Salmonella spp., we used characteristic colony morphologies, Gram staining, and additional tests, including sugar and motility tests. The strains were finally identified using API Listeria (bioMérieux, France).
3.9. Statistical Analysis
SPSS and Origin were used to analyze the experimental data and process the figures.
4. Results
4.1. Optimization of RT-LAMP Reaction Conditions
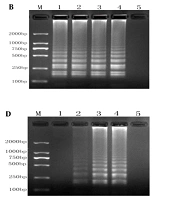
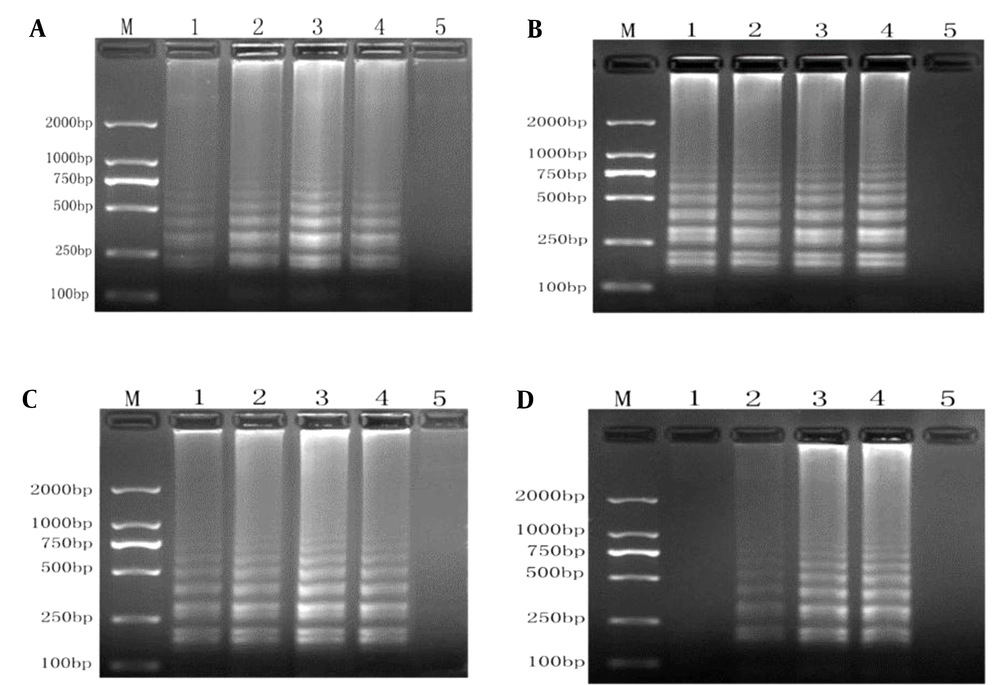
We optimized the RT-LAMP conditions by evaluating different reaction conditions. The optimal Mg and dNTP concentrations were 6.0 mM MgSO4 (Figure 2A) and 1.6 mM dNTPs (Figure 2C), respectively. Figure 2B shows a minor difference in temperature (63 and 65°C). No positive product was amplified at 40 U/mL Bst DNA polymerase, while the optimal results were obtained at 160 U/mL Bst DNA polymerase (Figure 2D). Accordingly, optimum conditions were 6.0 mM MgSO4, 1.6 mM dNTPs, and 160 U/mL Bst DNA polymerase at 65°C.
RT-LAMP system optimization diagram. A, optimization of Mg2+ concentrations (lane M, 2,000-bp ladder marker; lanes 1 - 4, the results of RT-LAMP with Mg2+ concentrations of 2.0 mmol/L, 4.0 mmol/L, 6.0 mmol/L, and 8.0 mmol/L; lane 5, negative control); B, optimization of annealing temperatures (lane M, 2000-bp ladder marker; lanes 1 - 4, the results of RT-LAMP at 59, 61, 63, and 65°C; lane 5, negative control); C, optimization of dNTP concentrations (lane M, 2000-bp ladder marker; lanes 1 - 4, the results of RT-LAMP with dNTP concentrations of 0.8 mmol/L, 1.2 mmol/L, 1.6 mmol/L, and 2.0 mmol/L; lane 5, negative control); D, optimization of Bst enzyme concentrations (lane M, 2,000-bp ladder marker; lanes 1-4, the results of RT-LAMP with Bst enzyme concentrations of 40, 80, 160, and 320 U/mL; lane 5, negative control).
4.2. Detection of RT-LAMP Products
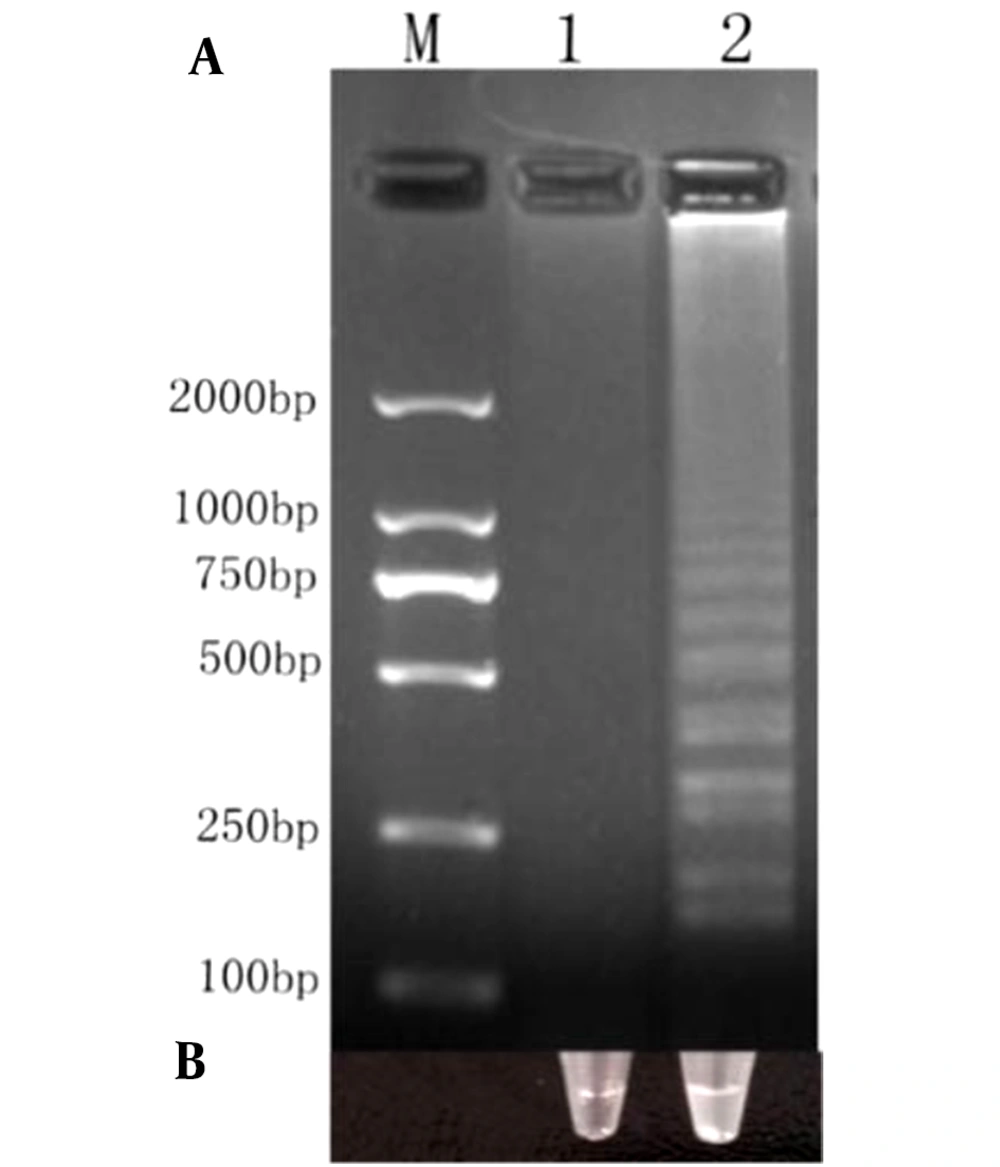
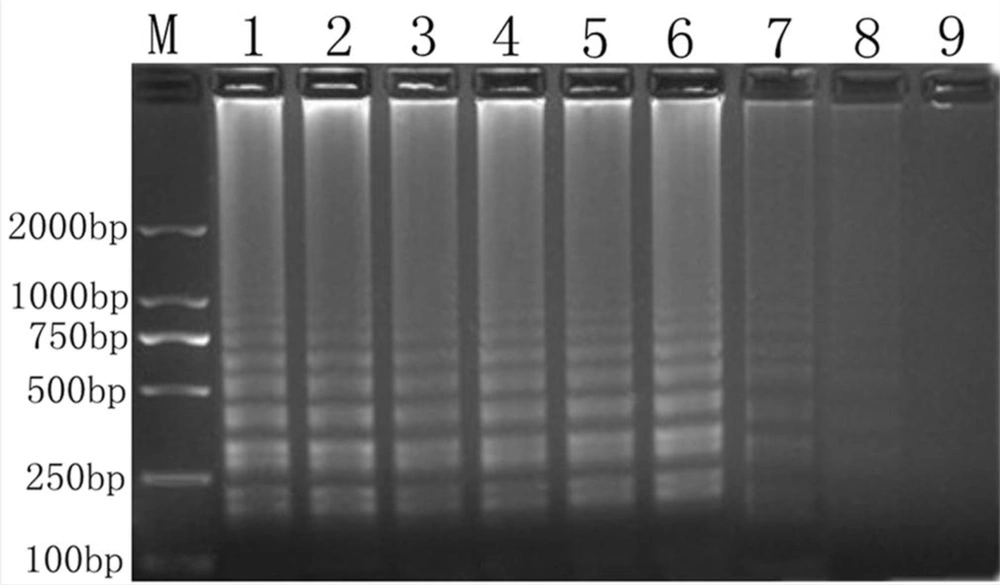
Our amplified products were separated by 2% agarose gel electrophoresis (Figure 3A), and the strains were observed with the naked eye (Figure 3B). We noted turbidity changes under normal lighting conditions. Positive cDNA samples were successfully amplified. The results revealed that Salmonella could be rapidly and easily detected using RT-LAMP.
4.3. Specificity of RT-LAMP Assay
Primer specificity (Figure 1) was investigated by extracting cDNA from eight strains of Salmonella and incorporating 33 strains of other species as templates (Table 1). Each of the eight Salmonella strains (Figure 4) was successfully amplified, whereas none of the other species were amplified. Accordingly, our primers were highly specific to Salmonella.
4.4. Sensitivity of RT-LAMP Assay
We performed sensitivity analysis by incorporating 10-fold serial dilutions of Salmonella, corresponding to 500 ng, 50 ng, 5 ng, 0.5 ng, 50 pg, 5 pg, 0.5 pg, and 50 fg (equivalent to 1 × 106 CFU/μL to 1 × 100 CFU/μL) of RNA. The RT-LAMP assay resulted in the DNA bands at all dilutions. However, the bands became weaker with decreasing RNA concentration. The sensitivity limit was 50 fg (Figure 5), equivalent to 1 CFU/μL.
4.5. Detection of viable Salmonella in RTE Food Samples
We assessed the capability of the RT-LAMP assay in detecting pathogenic microorganisms in the RTE foods, including a variety of vegetables, fruits, and meats (Table 3). The RT-LAMP and GB assays detected two Salmonella-positive chicken samples. Similar results were obtained for the other three types of food, indicating that the RT-LAMP and GB assays had similar detection capabilities.
| Sample (Tested Number) | No. of Samples Tested Positive for Salmonella | |
|---|---|---|
| RT-LAMP | GB 4789.4-2016 | |
| Beef (25) | 0 | 0 |
| Pork (25) | 1 | 1 |
| Egg (25) | 2 | 2 |
| Fish (25) | 0 | 0 |
| Total (100) | 3 | 3 |
Detection of Salmonella in RTE Foods Using RT-LAMP and GB 4789.4-2016 Assay
5. Discussion
Microbial contaminants have long been a health concern. Food poisoning has been studied for a long time, and plants are fully used as drugs in traditional treatment methods (18). In recent years, the abuse of antibiotics has led to the rapid spread of antibiotic resistance in pathogens (19) as such, it is necessary to change the traditional treatment strategy. Experts have conducted in-depth studies on the pathogenicity and defense mechanisms. Hassan Mahmoudi found out that the pathogenicity of Escherichia coli with iuc A and iut A genes may be associated to this issue (20). Propolis and garlic water extract can play a role in preventing pathogenic bacteria (21, 22). In the present study, a rapid detection method was designed for pathogenic bacteria in food to expand food safety methods further.
Developing detection methods for Salmonella, one of the most common contaminants in fruits and meats, is vital to minimize outbreaks (23). From a food safety and human health perspective, detecting foodborne bacterial pathogens requires immediate attention. This study proposed the use of RT-LAMP for detecting Salmonella in foods. The assay had high specificity and sensitivity in detecting Salmonella. Moreover, RT-LAMP was accurate and fast enough to be used in routine inspections and quarantine situations. Accordingly, it may serve as a potential diagnostic tool in the food industry to detect Salmonella in food products rapidly.
The proposed detection method of Salmonella uses inv A as the target gene, specific to Salmonella. Previous studies on inv A-based molecular detection assays have demonstrated their high specificity and sensitivity (24). When bacteria die, RNA cleaves quickly. Accordingly, the RT-LAMP assay for detecting Salmonella has lower false-positive results. Mg2+ concentration has the largest impact on primer annealing and DNA polymerase activity. A preliminary study revealed no target amplification at high Mg2+ concentrations since Mg2+ is bound to chelating agents such as ethylenediaminetetraacetic acid and anionic groups. Hence, the DNA polymerase activity and amplification results were affected. After evaluating a range of concentrations (2 - 8 mM), we found that 6 mM MgSO4 was optimal, indicating that the Bst DNA polymerase was effective at 61 - 65°C. We selected 65°C as the optimal temperature based on the AT and GC contents of the primers. The number of inv A mRNA copies was significantly higher than the number of inv A genes in the cell. Copy number is the major determinant of sensitivity.
5.1. Conclusions
The findings revealed that the detection limit for Salmonella in RT-LAMP was 1 CFU per reaction in pure cultures, which was 10-fold lower than that in PMA-LAMP (25). As an alternative to PCR, RT-LAMP is more sensitive, may be superior to PMA-LAMP in detecting Salmonella, and is further used to detect microorganisms in the RTE meats. Compared to GB 4789.4-2016, it was accurate; however, it took considerably less time.