1. Background
Staphylococcus aureus is normally found on the nasal mucosa and skin and may cause localized and severe invasive infections such as surgical wound infections, bacteremia, sepsis, toxic shock syndrome, and endocarditis (1, 2). Staphylococcus aureus has high propensity to develop antibiotic resistance (3). The emergence of methicillin-resistant Staphylococcus aureus (MRSA) in 1961 changed this bacterium into a superbug, which is now the most common multi-resistant organism (4) and accounts for a variety of difficult-to-treat infections in human (3). Methicillin-resistant S. aureus is usually divided into hospital-associated (HA-MRSA) and community-associated (CA-MRSA) groups (5). They differ in terms of their molecular characterization, antibiotic resistance pattern, pathogenicity, and virulence factors (6).
Community-associated-MRSA isolates are known because of their added virulence properties and highly transmissible nature with great potential to cause community-acquired diseases such as skin and soft tissue infections in healthy people with no defined predisposing factors (5). However, less commonly, similar to their hospital-acquired counterparts, CA-MRSA strains can cause rapidly fatal invasive infections, like bloodstream infection, septic shock necrotizing pneumonia, and necrotizing fasciitis (7, 8). Hospital associated-MRSA strains, in addition to β-lactam antibiotics are commonly resistant to many other antibiotics. In contrast, CA-MRSA strains are typically susceptible to multiple other anti-staphylococcal non-β-lactam antibiotics (6, 9, 10). Methicillin-resistant S. aureus isolates arises when methicillin-susceptible S. aureus (MSSA) acquires the mecA gene carried on the mobile Staphylococcal cassette chromosome mec (SCCmec) genetic element, which encodes an altered penicillin-binding protein (PBP2a) with a reduced affinity for β-lactam antibiotics (11).
Currently, at least 11 types of SCCmec element (I-XI) have been designated on the basis of their nucleotide sequence (7, 12). Commonly, it has been shown that the HA-MRSA isolates carry SCCmec types I, II, or III, while CA-MRSA isolates more often include SCCmec type IV, and less frequently, SCCmec types V or VII (11). However, previously, MRSA isolates harboring SCCmec type IV were isolated from hospital settings, and, vice versa, strains with SCCmec type III has been identified in the community setting, exhibiting the spread of these strains in both environments (6). The SCCmec IV cassette is small compared to other SCCmec types, and often lacks other antibiotic resistance genes beside mec A (7, 9). Another type of MRSA strain, as Oxacillin susceptible mecA-positive S. aureus (OS-MRSA) was emerged and is being steadily increasing worldwide (11). OS-MRSA isolates carry mec type gene but show oxacillin minimum inhibitory concentration (MIC) below the resistance breakpoints. These isolates generate highly methicillin-resistant subclones when exposed to β-lactam antibiotics (13). OS-MRSA isolates are misidentified as methicillin-sensitive using current phenotypic susceptibility tests, which may potentially cause treatment failure (13).
Asymptomatic S. aureus carriage is a risk factor for autogenous infections and cross-transmission to other individuals (14). The acquisition of antibiotic resistance in commensal bacteria is thought as a general threat to the public health. Methicillin-resistant S. aureus nasal carriage has shown a steady increase in the community setting during the time (15). Understanding the epidemiology of MRSA carriage and its antibiotic resistance properties in a community setting could help to design strategies to prevent the spread of the bacterium and empiric therapy of CA-MRSA infections (16).
2. Objectives
Since there are rare studies on MRSA nasal carriage rates in healthy Iranian children, this study was aimed to determine the prevalence and associated risk factors of nasal carriage, antibiotic resistance profile, and molecular characteristics of MRSA isolates in school children in Ardabil city, northwest Iran.
3. Methods
3.1. Subjects and Sampling
Subjects were students aged 12 - 14 years who were randomly selected from nine middle schools (male/female) in Ardabil city, located in Northwest of Iran. Between October to December 2017, 510 samples were taken from both nostrils of children using a sterile cotton swab. A questionnaire including variables like sex, age, weight, family size, number of students in the class, hospitalization in recent one year, hospitalization of family members in recent one year, antibiotic use in recent four weeks, antibiotic use in the past 12 months, ways of antibiotic usage (complete/incomplete course), the presence of health institution worker in the family, smoking, diabetes, nasal abnormalities, and acute watery diarrhea was filled out for each individual.
3.2. Bacterial Isolation and Identification
The swabs were first inoculated into the Tryptic Soy Broth culture medium (Merck, Darmstadt, Germany) and placed at 35 ± 2°C for 18 - 24 h. Then 10 µL of culture was subcultured onto Mannitol salt agar medium (BioMaxima, Lublin, Poland). The media were placed at 35 ± 2°C for 18 - 24 h. Staphylococcus aureus was characterized according to colony morphology, gram stain characteristics, catalase assay, DNase (Merck, Darmstadt, Germany) test, and tube coagulase test. Unequivocal identification of isolates as S. aureus was confirmed by detecting nuc gene (S. aureus thermonuclease gene), and for OR-MRSA strains was done by mecA gene (staphylococci methicillin resistance gene) using PCR test as reported earlier (17). Briefly, genomic DNA was extracted with a commercial DNA isolation kit (DNPTM, Sinaclon, Tehran, Iran) according to the manufacturer's recommendation.
Amplification was conducted in a DNA thermal cycler (BIO-RAD, California, USA) by temperature conditions, and specific primers are shown in Appendix 1 (18, 19). PCR products were confirmed by electrophoresis in a 1% agarose gel (Sinaclon, Tehran, Iran) and visualized by staining (DNA safe stain; Sinaclon, Tehran, Iran). Staphylococcus aureus ATCC 33591 genome and nuclease-free distilled water were used as positive (Sinaclon, Tehran, Iran) as the negative control respectively. Additionally, the representative PCR products were sequenced to verify the identity of the genes. Isolates were preserved at -80°C in Brain Heart Infusion broth (BioMaxima, Lublin, Poland), including 15% glycerol (Merck, Darmstadt, Germany) until further analysis.
3.3. Antimicrobial Susceptibility Testing
Antibiotic susceptibility testing was conducted by the disc diffusion method on Muller-Hinton agar (BioMaxima, Lublin, Poland) based on the guidelines outlined by Clinical and Laboratory Standards Institute (CLSI) (20). The tested antibiotics (Padtan Teb, Tehran, Iran) were penicillin (10 µg), co-amoxiclav (30 µg), chloramphenicol (30 µg), tetracycline (30 µg), ciprofloxacin (10 µg), ceftriaxone (100 µg), cefazolin (30 µg), clindamycin (2 µg), imipenem (10 µg), trimethoprim-sulfamethoxazole (25 µg), rifampicin (30 µg), gentamicin (10 µg) and mupirocin (5 µg). Phenotypic methicillin resistance was evaluated using oxacillin MIC testing (≥ 4 µg/mL indicated MRSA). The test was performed using agar dilution method according to CLSI instructions. Staphylococcus aureus ATCC 33591 was used as quality control strain (21, 22).
3.4. Molecular Typing
3.4.1. SCCmec Typing
SCCmec types (I, II, III, IVa, IVb, IVc, IVd, and V) were determined on all mecA-positive isolates using two series of multiplex-PCR assay as described previously with some modifications in annealing temperature and PCR program (20). The primer sequences and PCR conditions are listed in Appendix 1. PCR products were analyzed as outlined above. Representative PCR products were subjected to sequencing to confirm the identity of amplified genes.
3.4.2. Spa Typing Based Upon Repeat Pattern Analysis
The genetic profiles of the mecA-positive strains were also determined based on protein A (spa) typing. The polymorphic X region of spa gene was amplified using primers and PCR conditions as listed in Appendix 1. This method was previously described by Harmsen et al. (23). Nucleotide sequences of the amplified fragments were determined (Macrogen; Seoul, South Korea), and spa types were identified using online spa typing software (http://www.spaserver.ridom.de).
3.4.3. Enterobacterial Repetitive Intergenic Consensus (ERIC)-PCR
ERIC-PCR was conducted and analyzed, as previously described (24). Reactions were performed in a total volume of 50 µL containing 3.7 µL 10x buffer, 0.7 µL dNTP, 1 µL MgCl2, 2.5 µL of each primers ERIC1 and ERIC2 listed in Appendix 1, 0.5 µL Taq polymerase, 1 µL DNA template and 38.1 µL double depth water (DDW). ERIC-PCR was performed under the conditions are shown in Table 1. Amplicons were size-fractionated by agarose gel electrophoresis at 80 V for 2 h through 1.5% agarose gels, stained with DNA safe stain, and visualized and photographed as mentioned earlier. ERIC patterns were analyzed using GelQuest free trial version, v.3.4.3.0 (SequentiX – Digital DNA Processing, Germany). To ensure the validity of the results one representative isolate was included in every PCR testing set.
| Antibiotics | OR-MRSA (N = 31); No. (%) | OS-MRSA (N = 59); No. (%) | MSSA (N = 69); No. (%) | Total (N = 159); No. (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | I | R | S | I | R | S | I | R | S | I | R | |
| Penicillin | - | - | 31 (100) | - | - | 59 (100) | 3 (4.3) | - | 66 (95.6) | 3 (1.8) | - | 156 (98) |
| Amoxiclav | - | - | 31 (100) | - | - | 59 (100) | 65 (94.2) | - | 4 (5.7) | 65 (41) | - | 94 (59) |
| Chloramphenicol | 30 (96.7) | - | 1 (3) | 53 (89.8) | - | 6 (10.1) | 65 (94.2) | - | 4 (5.7) | 148 (93) | - | 11 (7) |
| Tetracycline | 16 (51.6) | 7 (22.5) | 8 (25.8) | 39 (66.1) | 7 (11.8) | 13 (22) | 56 (81) | 4 (5.7) | 9 (13) | 111 (70) | 18 (11) | 30 (19) |
| Ciprofloxacin | 29 (93.5) | - | 2 (6.4) | 53 (89.8) | 2 (3.3) | 4 (6.7) | 65 (94.2) | 1 (1.4) | 3 (4.3) | 147 (92.4) | 3 (1.8) | 9 (5.6) |
| Ceftriaxone | - | - | 31 (100) | - | - | 59 (100) | 65 (94.2) | 4 (5.7) | - | 65 (41) | 4 (2.5) | 90 (56) |
| Cefazolin | - | - | 31 (100) | - | - | 59 (100) | 66 (95.6) | 3 (4.3) | - | 66 (41.5) | 3 (1.8) | 90 (56) |
| Clindamycin | 18 (58) | 2 (6.4) | 11 (35) | 37 (62.7) | - | 22 (37.2) | 39 (56.5) | 2 (2.8) | 28 (40.6) | 94 (59) | 4 (2.5) | 61 (38) |
| Imipenem | - | - | 31 (100) | - | - | 59 (100) | 69 (100) | - | - | 69 (43) | - | 90 (56.6) |
| Rifampicin | 27 (87) | - | 4 (12.9) | 59 (100) | - | - | 69 (100) | - | - | 155 (97.4) | - | 4 (2.5) |
| Gentamycin a | 28 (90.3) | 1 (3.2) | 2 (6.4) | 57 (96.6) | 1 (1.6) | 1 (1.6) | 68 (98.5) | - | 1 (1.4) | 153 (96.2) | 2 (1.2) | 4 (2.5) |
| Mupirocin | 23 (74.1) | - | 8 (25.8) | 53 (89.8) | - | 6 (10.1) | 65 (94.2) | - | 4 (5.7) | 141 (88.6) | - | 18 (11) |
| Erythromycin a | 7 (22.5) | 2 (6.4) | 22 (71) | 15 (25.4) | 6 (10.1) | 38 (64.4) | 25 (36.2) | 6 (8.6) | 38 (55) | 47 (29.5) | 14 (9) | 98 (61.6) |
| Trimethoprim sulfamethoxazole | 29 (93.5) | - | 2 (6.4) | 54 (91.5) | 3 (5) | 2 (3.3) | 66 (95.6) | 2 (2.7) | 1 (1.4) | 149 (94) | 5 (3) | 5 (3) |
| Kanamycin a | 26 (83.8) | - | 5 (16.1) | 53 (89.8) | 1 (1.6) | 5 (8.4) | 59 (85.5) | 6 (8.6) | 4 (5.7) | 138 (87) | 7 (4.4) | 14 (9) |
| Tobramycin a | 6 (19.3) | 1 (3.2) | 5 (16.1) | 56 (94.9) | 1 (1.6) | 2 (3.3) | 67 (97) | 1 (1.4) | 1 (1.4) | 148 (93) | 3 (1.8) | 8 (5) |
| Vancomycin | 31 (100) | - | - | 59 (100) | - | - | 69 (100) | - | - | 159 (100) | - | - |
Antibiotic Susceptibility Profiles of OR-MRSA, OS-MRSA, and MSSA Strains Isolated from Nostrils of Healthy Children by Disk Diffusion Method in Ardabil, Northwest Iran
3.5. Detection of Virulence Genes
The presence of the genes encoding the staphylococcal enterotoxins (sea, seb, sec and sed), two exefoliative toxins (eta and etb), two hemolysin toxins (hla and hld), toxic shock syndrome toxin 1 (TSST-1) (tst), and the Panton-Valentine Leukocidin (PVL, lukF/S), was examined using PCR testing reported by others (9, 25).
3.6. Statistical Analysis
SPSS software, version 11.5, package for windows was used for the statistical analysis of the obtained data. Chi-square test was used to explore the underlying risk factors associated with MRSA nasal carriage. A P-value < 0.05 was considered statistically significant.
4. Results
4.1. Identification of Bacterial Isolates
A total of 159 S. aureus isolates were collected from 510 nasal samples obtained from the healthy children, including 233 males (45.7%) and 277 females (54.3%). Oxacillin MIC testing showed that 31 (19%) isolates were oxacillin resistant (Appendix 2), whereas PCR testing revealed in addition to 31 oxacillin resistant isolates (OR-MRSA), 59 (37.10%) oxacillin susceptible isolates were found to be mecA positive (OS-MRSA).
4.2. Antimicrobial Susceptibility Testing
The susceptibility patterns of all isolates, tested by disc diffusion method, are presented in Table 1. Overall, β-lactam antibiotics were the least active antibiotics against OR-MRSA and OS-MRSA isolates. Chloramphenicol (3%) and erythromycin (71%) were the most and least active non-β-lactam antibiotics against OR-MRSA isolates, respectively; Gentamycin (1.6%) and erythromycin (64.4%) were the most and least active non-β-lactam antibiotics against OS-MRSA isolates, respectively. Of β-lactam antibiotics, penicillin (95.6%) showed the least activity, and ceftriaxone (0%), cefazolin (0%), and imipenem (0%) showed the most activity against MSSA isolates. Gentamycin (1.4%), tobramycin (1.4%), and trimethoprim-sulfamethoxazole (1.4%) were the most active non-β-lactam antibiotics, whereas erythromycin (55%) showed the least active non-β-lactam antibiotic against MSSA isolates. Out of 31 OR-MRSA, 59 OS-MRSA, and 69 MSSA strains, 18 (58%), 29 (49%), and 29 (42%) isolates showed multidrug resistant phenotypes (resistant to 3 or more classes of antibiotics), respectively (Appendix 3).
4.3. SCCmec Typing
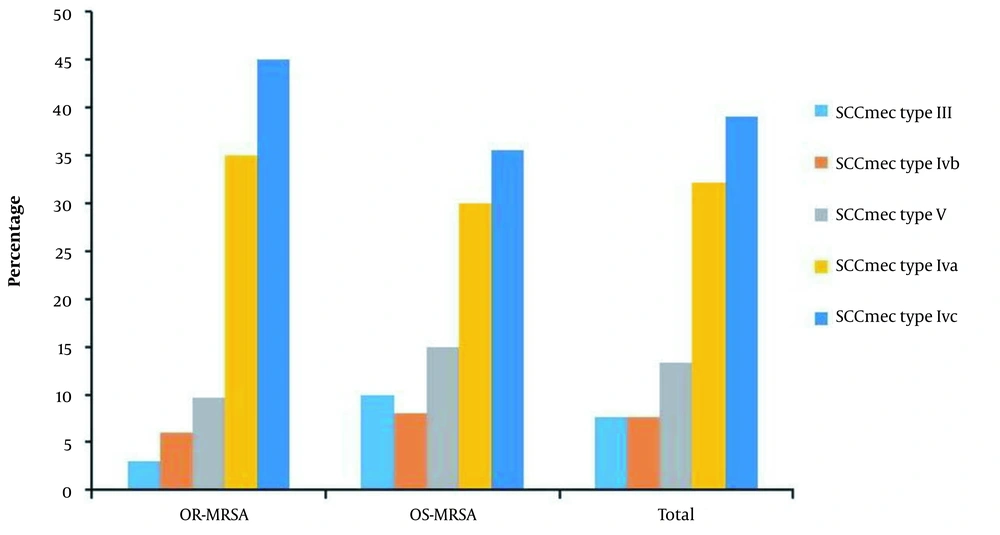
The SCCmec typing for OR-MRSA isolates showed that 14 (45%) out of 31 isolates were SCCmec type IVc, 11 (35%) isolates were SCCmec type IVa, 3 isolates (9.7%) were SCCmec type V, 2 (6.4%) isolates were SCCmec type IVb and a single isolate was SCCmec type III. The SCCmec typing for OS-MRSA isolates showed that out of 59 isolates, 21 (36%), 18 (30%), 9 (15%), 6 (10%) and 5 (8.5%) were SCCmec type IVc, SCCmec type IVa, SCCmec type V, SCCmec type III and SCCmec type IVb, respectively. SCCmec types I, II, and IVd were not observed (Figure 1).
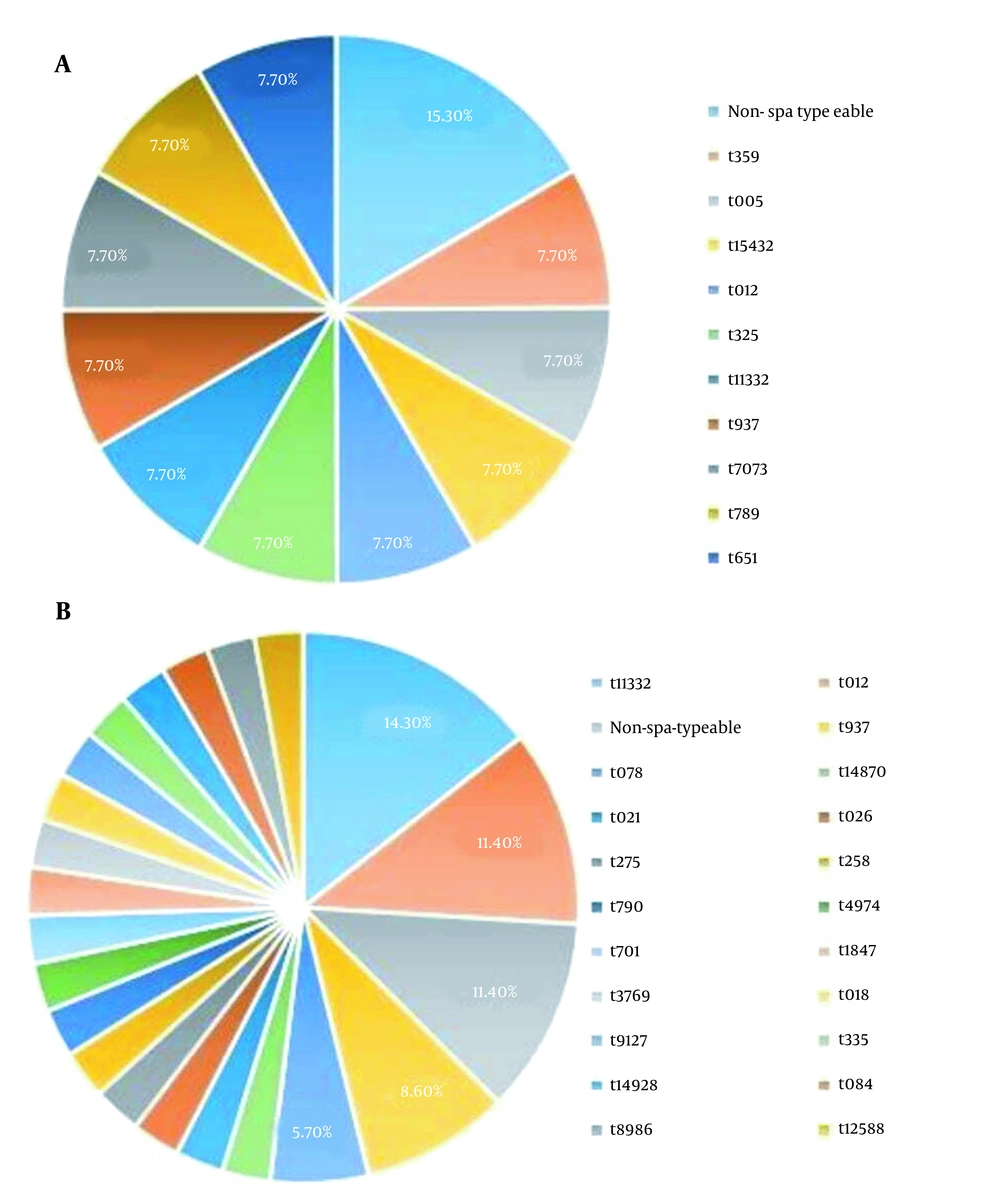
4.4. Spa Typing Based Upon Repeat Pattern Analysis
Eleven and 30 isolates were successfully spa typed in OR-MRSA, and OS-MRSA isolates, respectively. The results are shown in Figure 2. Collectively, 11 and 21 spa types were detected in OR-MRSA, and OS-MRSA isolates, respectively. The most common spa types were t11332 (14.3%) and t012 (11.4%) in OS-MRSA isolates. Of all spa types, only one type was identified in OR-MRSA isolates. Types t11332, t012, and t937 were found in both OR-MRSA, and OS-MRSA isolates.
4.5. ERIC-PCR Analysis
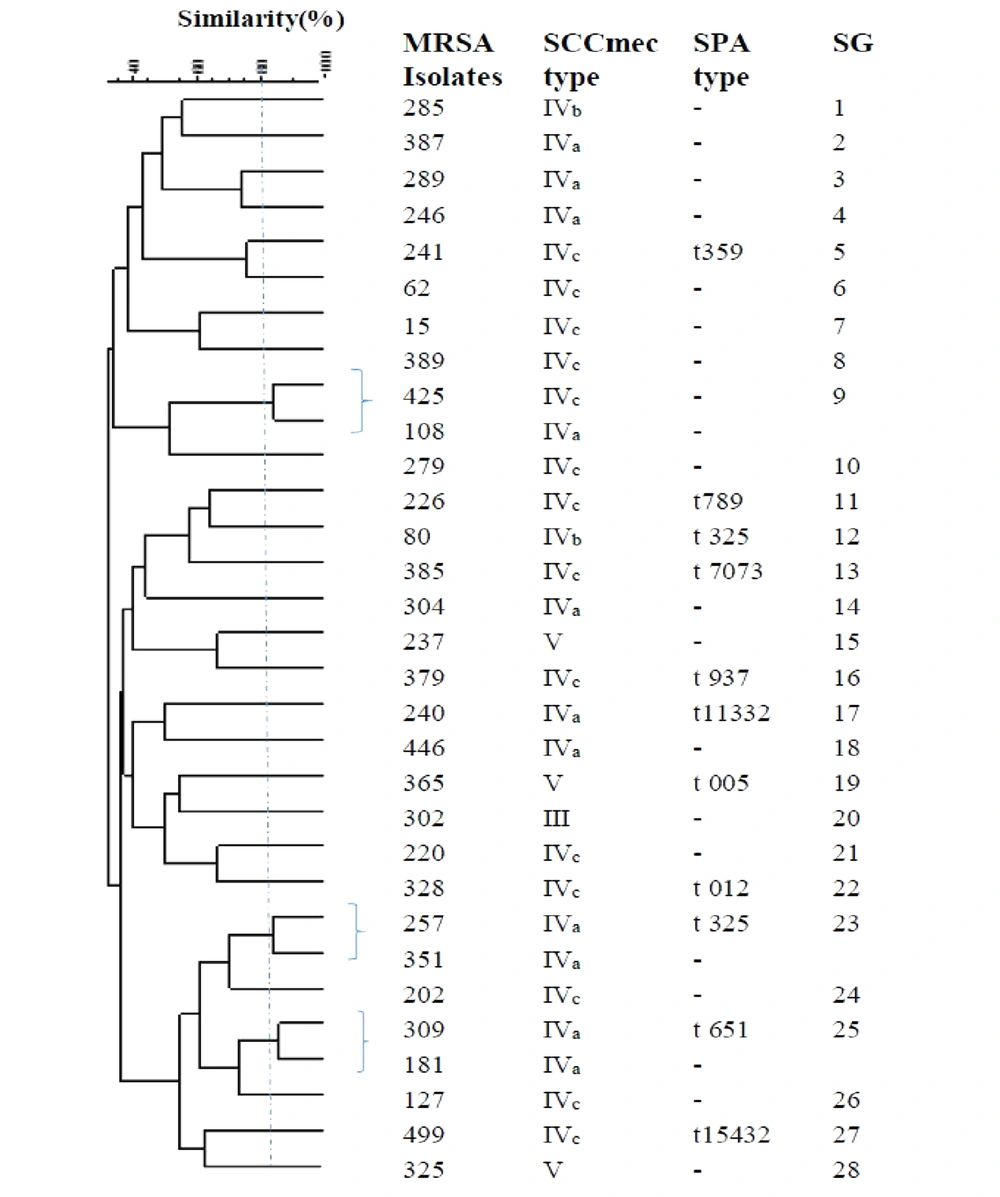
The ERIC-PCR primer in S. aureus generated 7 - 17 bands with molecular weights ranging from 50 to 1,500 bp. According to the band patterns with 80% similarity, the OR-MRSA and OS-MRSA isolates were classified into 28 and 47 different genotypes (subgroup), respectively (Figures 3 and 4). Of 31 OR-MRSA isolates and 59 OS-MRSA isolates tested, 25 and 39 isolates provided unique genotypes, respectively, whereas genotype subgroup 35 contained the highest number of isolates in OS-MRSA isolates (n = 4).
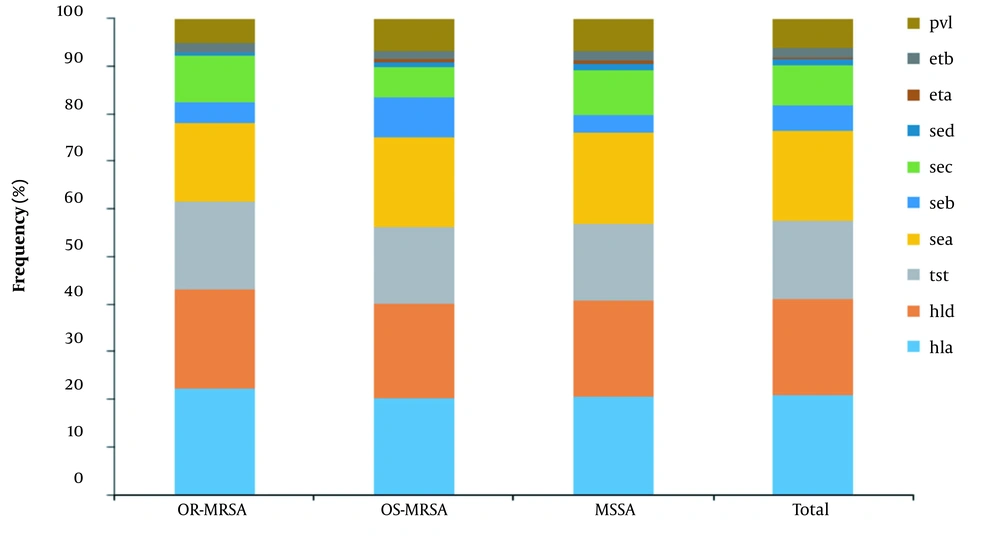
4.6. Detection of Virulence Genes
The distribution of virulence genes is shown in Figure 5. The hemolysin encoding hla (97%, 155/159) and hld (95%, 152/159) genes were the most frequent genes identified in isolates. The sea and sec with 139 (87%) and 63 isolates (40%) were the most common enterotoxin encoding genes followed by seb and sed with 42 (26%) and 8 (5%) isolates, respectively. The tst was found in 124 (78%) isolates, exfoliative toxins genes (eta and etb) were found in 4 (2.5%) and 14 (9%) isolates, respectively. The pvl gene was found in 46 (29%) isolates. In comparison, 25.50% of MRSA (9.60% OR-MRSA and 34% OS-MRSA) and 33% of MSSA isolates carried pvl gene. Appendix 4 shows the profile distribution of virulence genes in OR-MRSA, OS-MRSA, and MSSA isolates. A total of 47 different profiles were obtained, with profile sea, tst, hla, hld having the highest frequency. Overall, 97.48% of isolates carried ≥ 3 toxin encoding genes.
4.7. Identified Risk Factors
Regarding carriage status, totally, 31.17% (159/510) of subjects were S. aureus nasal carriers, including 13.50% (69/510), 6.07% (31/510), and 11.56% (59/510) as MSSA, OR-MRSA, and OS-MRSA carriers, respectively. Several risk factors were evaluated for possible association with OR-MRSA and OS-MRSA nasal carriage (Table 2). The number of students attending classrooms showed a significant association with carriage of OR-MRSA and OS-MRSA (P = 0.03). Previous antibiotic uptake (4 and 12 weeks before sample collection) was not associated with MRSA carriage (P > 0.05), while incomplete antibiotic course increased the risk of OS-MRSA carriage compared to OR-MRSA (P = 0.02) and MSSA (P = 0.001).
| Risk Factors | OR-MRSAN = 31, No. (%) | OS-MRSA N = 59, No. (%) | P-Value | OR-MRSA N = 31, No. (%) | MSSA N = 69, No. (%) | Resistance phenotypes | TotalmN = 159, No. (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| P-Value | OS-MRSA N = 59, No. (%) | MSSAnN = 69, No. (%) | P-Value | |||||||
| Age | 0.32 | 0.26 | 0.36 | |||||||
| 11 | - | - | - | 1 (1.4) | 1 (1.4) | 1 (0.6) | ||||
| 12 | - | 5 (8.5) | - | 8 (11.1) | 5 (8.5) | 8 (11.1) | 13 (8) | |||
| 13 | 12 (38.7) | 21 (35.6) | 12 (38.7) | 21 (30.4) | 21 (35.6) | 21 (30.4) | 54 (34) | |||
| 14 | 14 (45.2) | 25 (42.4) | 14 (45.2) | 22 (31.9) | 25 (42.4) | 22 (31.9) | 61 (38.3) | |||
| 15 | 4 (12.9) | 8 (13.6) | 4 (12.9 | 15 (22) | 8 (13.6) | 15 (22) | 27 (17) | |||
| 16 | 1 (3.2) | 1 (3.2) | 2 (2.8) | 2 (2.8) | 3 (1.9) | |||||
| Sex | 0.49 | 0.64 | 0.75 | |||||||
| Male | 15 (48.4) | 33 (55.9) | 15 (48.4) | 41 (59) | 33 (55.9) | 41 (59) | 89 (56) | |||
| Female | 16 (51.6) | 26 (44.1) | 16 (51.6) | 28 (40.6) | 26 (44.1) | 28 (40.6) | 70 (44) | |||
| Family size | 0.45 | 0.5 | 0.06 | |||||||
| Up to 4 | 22 (71) | 44 (74.6) | 22 (71) | 53 (77) | 44 (74.6) | 53 (77) | 119 (75) | |||
| > 4 to 10 | 9 (29) | 14 (23.7) | 9 (29) | 16 (23) | 14 (23.7) | 16 (23) | 39 (24.5) | |||
| > 10 | - | 1 (1.7) | - | - | 1 (1.7) | - | 1 (0.6) | |||
| Number of children in classroom | 0.03a | 0.2 | 0.07 | |||||||
| 23 to 30 | 24 (77.5) | 51 (86.5) | 24 (77.5) | 52 (72.2) | 51 (86.5) | 52 (72.2) | 127 (78.5) | |||
| > 30 to 37 | 7 (22.6) | 8 (13.6) | 7 (22.6) | 17 (25) | 8 (13.6) | 17 (25) | 32 (20) | |||
| Hospital admission in past 1 year | 2 (6.5) | 3 (5.1) | 0.78 | 2 (6.5) | 7 (58.3) | 0.56 | 3 (5.1) | 7 (58.3) | 0.31 | 12 (7.3) |
| Hospitalization of family members in past 1 year | 10 (32.3) | 11 (18.6) | 0.14 | 10 (32.3) | 15 (22) | 0.34 | 11 (18.6) | 15 (22) | 0.44 | 36 (23) |
| Taking antibiotic in past 4 weeks | 10 (32.3) | 18 (30.5) | 0.86 | 10 (32.3) | 23 (33.3) | 0.80 | 18 (30.5) | 23 (33.3) | 0.60 | 51 (32) |
| Taking antibiotic in past 12 months | 18 (58.1) | 28 (47.5) | 0.33 | 18 (58.1) | 37 (54) | 0.7 | 28 (47.5) | 37 (54) | 0.42 | 83 (52) |
| Ways of antibiotic usage | 0.02 a | 0.56 | 0.001 a | |||||||
| Complete course | 15 (48.4) | 15 (25.4) | 15 (48.4) | 38 (55 ) | 15 (25.4) | 38 (55) | 68 (43) | |||
| Incomplete course | 16 (51.6) | 44 (74.6) | 16 (51.6) | 31 (45) | 44 (74.6) | 31 (45) | 91 (57) | |||
| Presence of health institution worker in the family | 4 (12.95) | 7 (11.9) | 0.88 | 4 (12.95) | 5 (6.9) | 0.31 | 7 (11.9) | 5 (6.9) | 0.32 | 16 (9.85) |
| Smoking | - | 2 (3.4) | 0.30 | - | - | - | 2 (3.4) | - | 0.11 | 2 (1.2) |
| Diabetes | - | 1 (1.7) | 0.46 | - | - | - | 1 (1.7) | - | 0.26 | 1 (0.6) |
| Nasal abnormalities | 3 (9.7) | 3 (5.1) | 0.40 | 3 (9.7) | 6 (9 ) | 0.95 | 3 (5.1) | 6 (9) | 0.31 | 12 (7.5) |
| Having diarrhea | - | - | - | - | 1 (1.4) | 0.5 | - | 1 (1.4) | 0.36 | 1 (0.6) |
Factors Associated with OR-MRSA, OS-MRSA and MSSA Carriage in Healthy Children in Ardabil, Northwest Iran
5. Discussion
Staphylococcus aureus has been recognized as a causative agent of human infections since the late 1870s. However, its role as a colonizer of healthy people was taken into consideration more than 50 years later (14). Although there is a large amount of information on the frequency and significance of the healthy carrier state of S. aureus in the literature (14), studying on its epidemiology is still continuing due to dynamic changes in the genome of the bacterium by acquisition of new antibiotic resistance genetic elements and its increasing role in both nosocomial and community-associated infections (14). Previous cross-sectional studies indicated approximately 25-30% community carriage rates for S. aureus around the world (26). In the present study, 159 out of 510 individuals screened, were S. aureus nasal carriers (31.2%), which is above the upper limits of the community carriage rates mentioned above. Similar findings as 31, 35, 37, and 47.7% carriage rates were reported in children from Nepal, India, Turkey, and Ethiopia, respectively (3, 8, 27, 28).
Biologically, S. aureus carriage rate is dependent on receptors on host epithelial cells and bacterium adhesive factors (29). However, higher community carriage rates for S. aureus in our study could be attributed to some extent to the age group of the subjects recruited. Some studies earlier showed the high proportions of colonization and transient carriage rate in children of young age groups (26, 29). Nasal carriage of S. aureus by itself is important as a risk factor for later infection and cross-transmission to other individuals. However, colonization with MRSA strains is much more important since infections with these strains are usually difficult to treat and associated with higher mortality rates (8). Screening and decolonization of MRSA nasal carriage on hospital admission is an effective infection control measure and reduces the subsequent staphylococcal infections (4).
In the present study, 90 out of 510 (17.5%) individuals were colonized with mecA positive S. aureus strains (6.07% OR-MRSA and 11.56% OS-MRSA). Most previous reports on community MRSA carriage include OR-MRSA isolates. In a systematic review performed from 2000 to 2016 in Asian pacific countries, a carriage rate of 0 to 23.5% was reported for CA-MRSA among the healthy population. India (16.5 - 23.5%), followed by Vietnam (7.9%), and Taiwan (3.5 - 3.8%) were on the top list of the countries with high CA-MRSA carriage prevalence (30). Compared to the above-mentioned information, the community level prevalence of OR-MRSA carriage is relatively high in our population. Lack of immunity to CA-MRSA and close contact with asymptomatic CA-MRSA carriers may increase colonization of MRSA among young children (30). The term OS-MRSA for the description of S. aureus was first used in Japan (31). A few studies have characterized the resistance mechanisms in-depth in several OS-MRSA isolates (32).
In the present study, 11.56% of children were OS-MRSA nasal carriers. There are limited data on OS-MRSA community carriage to compare the results. However, the pieces of evidence show the growing trend of OS-MRSA carriage in Iran. In 2013 in a study performed by authors on nursing staff in a teaching hospital, 2 out of 173 (1.15%) cases were OS-MRSA carriers (22). Another study carried by Zeinalpour Ahrabi et al. in 2018 on high school students in East Azerbaijan province in Iran revealed 6.25% of the students were OS-MRSA carriers (33). In a study carried out in some African countries, 2.4% of healthcare workers were found to be OS-MRSA carrier (30). Despite the lack of the studies on OS-MRSA carriage status, for over a decade, high prevalence of clinical, livestock, and environmental OS-MRSA isolates have been reported from various countries in the world (11, 34, 35). Transmission of OS-MRSA is a matter of concern because the bacterium is not phenotypically resistant to oxacillin but can become high-level β-lactam resistant upon exposure to antibiotics during therapy, which may result in treatment failure (31).
The genetic diversity of OS-MRSA and OR-MRSA isolates was studied using SCCmec, spa, and ERIC-PCR typing methods. The SCCmec element represents the distinction between HA-MRSA and CA-MRSA strains (11). In the present study, the most common SCCmec types in OR-MRSA and OS-MRSA isolates were type IV with 80 and 71% followed by type V with 9.7 and 10%, respectively. These findings are consistent with previous reports showing that CA-MRSA mostly carries SCCmec types IV and V, whereas HA-MRSA usually possesses SCCmec types I, II, and III (6, 10, 12, 36). However, there are studies showing the intrusion of HA-MRSA into the community and CA-MRSA into the healthcare settings (6, 10, 36). In our study, 3.1% of OR-MRSA and 10% of OS-MRSA isolates carried SCCmec type III. This finding clearly points to the circulation of clones of MRSA in our community, which carry SCCmec elements from the hospital as well.
Studying the nucleotide sequence of the variable repeat region, the staphylococcal protein A encoding gene (spa typing) offers a rapid and accurate test to differentiate S. aureus outbreak isolates (37). Previously, the spa types of t437, t020, t064, t084, t242, t037, t030, t002, t008, and t032 were reported as the10 most common types in S. aureus clinical isolates worldwide (38). In Asian countries, spa types t030, t037, and t002 were the most prevalent types, with t037 as the predominant type in Iran (38). In the present study, significantly divergent and distinct spa type pattern was observed among our isolates. Eleven OR-MRSA and 30 OS-MRSA isolates were evaluated using spa typing method in which 11 and 21 different types were observed, respectively. The types t11332 (14.3%) and t012 (11.4%) were the most prevalent ones found in OS-MRSA isolates. Most prevalent circulating spa type in Iran, t037 was not observed among our isolates. In a similar study conducted on OR-MRSA isolates collected from healthy children in Iran, the spa types t790, t267, and t2962 were found to be the most common types, and the spat type11332 were identified in 3.83% of isolates (39).
Staphylococcus aureus is a forerunner bacterium in acquisition of resistance genes. Antibiotic resistance is the main concern in the treatment of staphylococcal infections. In the present study, overall, 48% of isolates were MDR, which is higher than the rates previously reported in isolates recovered from healthy children in other Iranian cities, including Kashan (2014) and Tabriz (2019), with 29.3 and 31.66% MDR prevalence, respectively (33, 40). The prevalence of MDR isolates was higher in OR-MRSA (58%), and OS-MRSA (49%) strains in comparison to MSSA (42%) isolates. For most tested antibiotics, the resistance rate was significantly higher in OR-MRSA compared to OS-MRSA and MSSA isolates.
Excluding β-lactam antibiotics, it has been documented that CA-MRSA resistance is high for erythromycin, tetracycline, clindamycin, co-trimoxazole, and gentamicin (30). Similarly, high rates of resistance was observed for erythromycin (71%), tetracycline (25.8%), clindamycin (35%) in our isolates. In contrast, the resistance rate was not high against co-trimoxazole and gentamicin in our isolates. The prevalence of antibiotic-resistant bacteria colonization among healthy people is different depending on geographical regions. Complex socioeconomic and behavioral factors affect the emergence and distribution of antibiotic-resistant bacteria (41). High rates of resistance to erythromycin in our CA-MRSA isolates suggest the limited possibility for erythromycin to be used in treatment of CA-MRSA infections. Resistance toward tetracycline and clindamycin are also a matter of concern, because they have been proposed for treatment of MRSA SSTIs (30). Additionally, clindamycin is even being prescribed for treatment of pneumonia, joint, and bone infections caused by MRSA (30).
Surprisingly, 11% of our isolates [OR-MRSA (25.8%), OS-MRSA (10.1%), and MSSA (5.7%) isolates] were found to be resistant to mupirocin. Previously it has been shown the absence of mupirocin resistance among S. aureus isolates from nasal carriers in general population in community setting in Asia-Pacific countries (30). Emergence of mupirocin resistance among carriage isolates is of great concern. Because mupirocin is used for decolonization of MRSA in the nose of certain groups of patients at hospital admission (42). Toxins play a crucial role in promoting staphylococcal infections (1). In this study, the majority of isolates were carried multiple toxins encoding genes. Overall, ≥ 3 toxin encoding genes were detected in 97.48% of isolates. In the current study, hla and hld were the most frequent virulence genes detected. These genes encode the α-hemolysin (also known as α-toxin) and δ-hemolysin (also named δ-toxin), respectively.
The hemolysins are cytolytic agents with the ability to damage a wide range of host cells, including neutrophils, monocytes, and macrophages, and can significantly contribute to inhibiting both innate and adaptive immune responses against S. aureus infection (43). Moreover, α-hemolysin is among the main staphylococcal toxins inducing pathological injury, and δ-toxin is part of the agr (accessory gene regulator) locus that contributes to control of other staphylococcal enzymes and toxins production (44). In addition, α-hemolysin is encoded in the core-genome, while others are encoded by acquired mobile genetic elements (44). Consistent with our results, a report by Yu et al. from china showed almost all of the isolates contained hla and hld (43). However, opposite to our study, no isolate was found to be positive for hld in other reports from China and Japan (6, 31, 45).
Panton-Valentine leukocidin (PVL) is a pore-forming toxin encoded by two co-transcribed genes (lukF-PV and lukS-PV) of a prophage inserted in the S. aureus chromosome. The importance of PVL as a potential virulence factor has been recognized. MRSA isolates producing PVL toxin are often responsible for severe skin infections. Worldwide, it has been shown that carrying of pvl gene is high in CA- MRSA compared to HA-MRSA isolates (31). In our study, 33 and 25.50% of MRSA and MSSA isolates carried pvl gene, respectively. It was significantly higher in OS-MRSA (34%) compared to OR-MRSA (9.60%) isolates. While low frequent occurrence (0 - 3.4%) for pvl gene has been reported in OS-MRSA isolates previously (31, 46). Staphylococcal enterotoxins are the cause of staphylococcal food poisoning. SEA and SEB are ranked as the first and second most common enterotoxins responsible for staphylococcal food poisoning throughout the world (1). According to a study carried out in Turkey, SEA was found to be the most common enterotoxin (40.1%) in hospital and community-acquired S. aureus isolates (47). Consistent with previous reports, sea gene was the most predominant SE encoding gene (47) identified in the current study, followed by sec (40%), seb (26%), and sed (5%).
Staphylococcus aureus isolates producing TSST-1 are usually associated with complicated diseases. The tst gene was detected in 78% of isolates in our study. Other studies around the world reported lower frequent occurrence (1.5 - 39%) for tst gene previously (1, 6, 31, 33). The exfoliative toxins (encoded by eta and etb) are responsible for staphylococcal scaled-skin syndrome (SSSS). The global exfoliative toxin genes occurrence has been reported in up to 5% of S. aureus human isolates (48, 49). In our study, 4 (2.5%) and 14 (9%) isolates carried the eta and etb gene, respectively. However, previous studies reported higher occurrence (34 - 61%) for these genes in colonizing and clinical S. aureus isolates (7, 31, 50). Several risk factors have been found to be associated with MRSA nasal carriage (29). The human carriers are the most important source for the transmission of S. aureus. It has been shown that living in a crowded area and large families increase the risk of S. aureus nasal carriage (29). This association is probably because of the increased sharing of nasal flora within a large community. In this study, in classrooms with 23 to 30 children prevalence of OR-MRSA (77.5%) and OS-MRSA (86.5%) was significantly high.
A similar study by Kejela and Bacha (2013) indicated that the number of children per classroom was significantly increased the S. aureus and MRSA nasal carriage (3). In contrast to the number of students in classrooms, there was no association between family size and MRSA carriage in this study. This may be because of the lower average of household members (75%; ≥ 4 member) in this study. In a similar study, family size of more than 10 members was independently associated with nasal carriage of S. aureus (8). It has been previously shown that health care workers are at greater risk of colonizing with MRSA isolates (22) and subsequently transmitting the bacteria to their family members (51). However, the results of this study did not show a significant association between MRSA carriage and the presence of health institution worker in the family. This result may be due to the fact that the number of families with a health institution employee was too small. In the literature, previous use of antibiotic has been described as a risk factor for MRSA carriage in children (52). However, in the present study, the history of antibiotic uptake was not associated with MRSA carriage, while the incomplete antibiotic course was identified as a strong risk factor for OS-MRSA nasal carriage in comparison to OR-MRSA and MSSA. Previous studies have shown a clear association between exposure to antibiotics and MRSA isolation (53).
5.1. Conclusions
This study indicated a high proportion of multi-resistant CA-MRSA nasal carriage in an Iranian healthy school children community. Carriage of OS-MRSA isolates is of greater concern as this phenotype cannot be detected using conventional laboratory methods, and treatment of OS-MRSA infections with β-lactam antibiotics may result in the emergence of high-resistant OR-MRSA and treatment failure. The results of this study provided an important insight into the status of S. aureus carriage in children and could be useful in the establishment of effective infection control measures to stop the dissemination of the bacterium.