1. Background
Pseudomonas aeruginosa is a small Gram-negative bacillus that is commonly arranged in pairs. It is an obligately aerobic, non-fermentative, and oxidase-positive bacterium (1-3). This opportunistic microorganism is ubiquitous, especially in hospitals, and is rarely related to infections in the natural host. Nevertheless, P. aeruginosa is a causative agent for 8% of all hospital-acquired infections, including pulmonary infections (tracheobronchitis and necrotizing bronchopneumonia), skin and soft-tissue infections (burn wounds and surgical site infections), urinary tract infections, bacteremia, and endocarditis (1, 4). Acute and chronic infections of P. aeruginosa are more common in hospitalized patients in the intensive care unit (ICU), ventilator-dependent and immunocompromised patients, and those receiving broad-spectrum antibiotics (1, 4). On the other hand, there are community-acquired infections associated with P. aeruginosa, including ulcerative keratitis, otitis externa, skin, and soft-tissue infections (diabetic foot infections) (5).
Simple growth requirements, broad environmental distribution, antimicrobial resistance, and virulence factors account for the widespread P. aeruginosa infections (1, 6). Pseudomonas aeruginosa drug resistance and the presence of virulence factors have been shown in numerous studies. However, there are no data on the prevalence of P. aeruginosa antibiotic resistance and virulence factors profile in Ardabil city of Iran. Antibiotic resistance in P. aeruginosa strains has an intrinsic, acquired, or adaptive characteristic (1). Based on the Centers for Disease Control and Prevention (CDC) report, multidrug-resistant (MDR) P. aeruginosa strains are a significant threat to public health, and are associated with 13% of hospital-acquired infections (4). On the other hand, drug-resistant and virulent strains of P. aeruginosa possess many virulence factors such as adhesins (flagella, pili, lipopolysaccharide (LPS), and alginate), secreted toxins (exotoxin A), enzymes (elastases, phospholipases, and exoenzymes), and secretion systems, which contribute to the development of severe infections that are hard to treat (1, 7).
The most common P. aeruginosa virulence factors that are assessed in the current study include: (1) pili, encoded by the pilB gene; (2) alginate, encoded by the algD gene, which is an exopolysaccharide capsule with an important role in bacterial survival against host's immune response and drugs; (3) sialidase, encoded by the nan1 gene, which is involved in the adherence to the respiratory tract; (4) exotoxin A, encoded by the toxA gene, which disrupts protein synthesis in eukaryotic cells; (5) las B elastase, a zinc metalloprotease encoded by the lasB gene, which destroys the collagen and elastin proteins in lung tissue; (6) hemolytic phospholipase C, encoded by the plcH gene; (7) non-hemolytic phospholipase C, encoded by the plcN gene; (8) exoenzyme S, encoded by the exoS gene, which is involved in colonization, invasion, and bacterial dissemination; and (9) exoenzyme U, encoded by the exoU gene, which is involved in tissue destruction and inflammatory response (1, 8).
2. Objectives
Awareness of the distribution of virulence genes and antibiotic resistance in clinical isolates is important for understanding P. aeruginosa infections epidemiology. Therefore, this study aimed to assess the P. aeruginosa drug resistance pattern and the most prevalent P. aeruginosa virulence gene profiles.
3. Methods
3.1. Pseudomonas aeruginosa Strains
Eighty-four clinical isolates of P. aeruginosa were collected from five hospitals (Imam Reza, Imam Khomeini, Bu-Ali, Alavi, Sabalan, Fatemi, and Ghaem) in Ardabil, Iran, between June 2019 and February 2021. Pseudomonas aeruginosa isolates were obtained from various specimens including wound, blood, urine, sputum, and cerebrospinal fluid (CSF) and then identified using standard microbiology tests such as phenotypic methods, i.e., pigment production, Gram staining, colony morphology, and oxidase and the genotypic method using PCR with specific primers of species (9). Isolated bacteria were then stored in cryovials containing tryptic soy broth plus 20% glycerol at -20°C until drug susceptibility testing, genomic DNA extraction, and PCR assay.
3.2. Drug Susceptibility Testing
Pseudomonas aeruginosa susceptibility patterns were determined using the Kirby-Bauer disk diffusion method against three antibiotic groups recommended by the Clinical and Laboratory Standards Institute (CLSI) guideline (10). They were group A antibiotics including piperacillin-tazobactam (100/10 μg), ceftazidime (30 μg), gentamicin (10 μg), and tobramycin (10 μg), group B antibiotics including cefepime (30 μg), aztreonam (30 μg), imipenem (10 μg), meropenem (10 μg), doripenem (10 μg), amikacin (30 μg), ciprofloxacin (5 μg), and levofloxacin (5 μg), and group O antibiotics including piperacillin (100 μg), ticarcillin-clavulanate (75/10 μg), netilmicin (30 μg), norfloxacin (10 μg), lomefloxacin (10 μg), and ofloxacin (5 μg) (Padtan Teb, Iran, Cypress Diagnostics, Belgium). For this purpose, a bacterial standard suspension (0.5 McFarland turbidity standard) equivalent to 1.5 × 108 CFU/mL was prepared and then poured on Mueller-Hinton agar medium (Conda, Pronasida, Spain). Antibiotic disks with specific concentrations were placed onto the lawns of bacteria and then incubated for 16 - 18 hours at 37°C. Inhibition zones around each disk were measured based on the CLSI guidelines and reported as susceptible, intermediate, or resistant. Pseudomonas aeruginosa ATCC 27853 was used for quality control of disks, media, inoculum preparation, and zones’ measurement in drug susceptibility tests.
3.3. Genomic DNA Extraction and PCR Assay
Bacterial genomic DNA from each confirmed P. aeruginosa strain was extracted using the boiling method and used for amplifying the selected virulence genes by an Eppendorf Thermal Cycler (Hamburg, Germany). Table 1 shows the oligonucleotide primers (SinaClon, Iran) along with programs used in the PCR assay. Each reaction of PCR assay was performed in a volume of 25 μL containing 20 μL of master mix (Tris-HCl pH 8.5, (NH4)2SO4, 3 mM MgCl2, 0.2% Tween 20, 0.4 mM dNTPs, 0.2 units/μL Ampliqon Taq DNA polymerase) (Ampliqon, Denmark), 1 μL of each of the forward and reverse primers (10 μmol/L), and 3 μL of template DNA. The presence of P. aeruginosa virulence genes in PCR products was detected using 1% agarose gel electrophoresis, with the TBE 0.5x buffer (Tris-Borate-EDTA) at 100 V for one hour, and then confirmed by sequencing.
| Gene | Oligonucleotide Sequence (5′ to 3′) | Thermal Cycling Condition for Amplification | Amplicon Size (bp) | References |
|---|---|---|---|---|
| algD | F: CGTCTGCCGCGAGATCGGCT; R: GACCTCGACGGTCTTGCGGA | Initial denaturation at 95°C for 5 min (1 cycle); Denaturation at 94°C for 1 min, annealing at 63°C for 1.5 min; extension at 72°C for 1 min (30 cycles) | 313 | (11) |
| lasB | F: GGAATGAACGAAGCGTTCTCCGAC; R: TTGGCGTCGACGAACACCTCG | Initial denaturation at 95°C for 5 min (1 cycle); Denaturation at 94°C for 1 min, annealing at 63°C for 1.5 min, extension at 72°C for 1 min (30 cycles) | 284 | (11) |
| plcH | F: GCACGTGGTCATCCTGATGC; R: TCCGTAGGCGTCGACGTAC | Initial denaturation at 95°C for 5 min (1 cycle); Denaturation at 94°C for 1 min, annealing at 60°C for 1.5 min, extension at 72°C for 1 min (30 cycles) | 608 | (11) |
| plcN | F: TCCGTTATCGCAACCAGCCCTACG; R: TCGCTGTCGAGCAGGTCGAAC | Initial denaturation at 95°C for 5 min (1 cycle); Denaturation at 94°C for 1 min, annealing at 63°C for 1.5 min, extension at 72°C for 1 min (30 cycles) | 481 | (11) |
| exoU | F: GGGAATACTTTCCGGGAAGTT; R: CGATCTCGCTGCTAATGTGTT | Initial denaturation at 95°C for 5 min (1 cycle); Denaturation at 94°C for 1 min, annealing at 56°C for 1.5 min, extension at 72°C for 1 min (30 cycles) | 428 | (12) |
| exoS | F: CGTCGTGTTCAAGCAGATGGTGCTG; R: CCGAACCGCTTCACCAGGC | Initial denaturation at 95°C for 5 min (1 cycle); Denaturation at 94°C for 1 min, annealing at 60°C for 1.5 min, extension at 72°C for 1 min (30 cycles) | 444 | (11) |
| toxA | F: CTGCGCGGGTCTATGTGCC; R: GATGCTGGACGGGTCGAG | Initial denaturation at 95°C for 5 min (1 cycle); Denaturation at 94°C for 1 min, annealing at 63°C for 1.5 min, extension at 72°C for 1 min (30 cycles) | 270 | (11) |
| nan1 | F: AGGATGAATACTTATTTTGAT; R: TCACTAAATCCATCTCTGACCCGATA | Initial denaturation at 95°C for 5 min (1 cycle); Denaturation at 94°C for 1 min, annealing at 49°C for 1 min, extension at 72°C for 1 min (30 cycles) | 1316 | (13) |
| pilB | F: ATGAACGACAGCATCCAACT; R: GGGTGTTGACGCGAAAGTCGAT | Initial denaturation at 95°C for 5 min (1 cycle); Denaturation at 94°C for 1 min, annealing at 57°C for 1 min, extension at 72°C for 1 min (30 cycles) | 826 | (12) |
3.4. Statistical Analysis
The prevalence of virulence genes and antibiotic resistance profile based on gender, sample source, and hospital ward was determined and then compared between different groups using the chi-square test. A P value of < 0.05 was considered statistically significant. All analyses were performed using SPSS software (version 16).
4. Results
4.1. Characteristics of the Patients and Specimens
A total of 84 P. aeruginosa strains were isolated from different clinical specimens in this cross-sectional study. The detailed information of the patients, specimens, and hospital wards were as follows: (1) 47.6% (n = 40) of P. aeruginosa strains were obtained from males and 52.4% (n = 44) from females. The mean age of the patients was 56.6 ± 18.6 years (6 - 89); (2) 46.5% (n = 39) of P. aeruginosa strains were obtained from Alavi hospital, 32.1% (n = 27) from Imam Khomeini hospital, 10.7% (n = 9) from Imam Reza hospital, 7.1% (n = 6) from Bu-Ali hospital, and 3.6% (n = 3) from Sabalan, Fatemi and Ghaem hospitals; (3) 51.2% of P. aeruginosa strains were isolated from urine specimens (n = 43), 17.9% from blood (n = 15), 19% from sputum (n = 16), 10.7% from wound (n = 9), and 1.2% (n = 1) from CSF; (4) The highest rate of P. aeruginosa strains isolated belonged to the ICU (32.1%, n = 27), followed by internal (31%, n = 26), emergency (16.6%, n = 14), neurology (17.9%, n = 15), and pediatric (2.4%, n = 2) wards.
4.2. Antibiotic Resistance Profile
The distribution of antibiotic resistance profiles is shown in Table 2. The highest and the lowest antibiotic resistance rates of P. aeruginosa were against ticarcillin-clavulanate (94%) and doripenem (33.3%), respectively. In addition, the prevalence of MDR isolates was 55.9% (n = 47). Pseudomonas aeruginosa isolates resistant to at least one antibiotic in the three classes of antibiotics were considered as MDR strains (9). The presence of statistically significant associations between the prevalence of antibiotic resistance and virulence genes was as follows: piperacillin vs. exoU and pilB, piperacillin-tazobactam vs. exoU and pilB, ticarcillin-clavulanate vs. exoU, ceftazidime vs. exoU and pilB, cefepime vs. exoU and pilB, aztreonam vs. plcH, plcN, and pilB, doripenem vs. exoU, nan1, and pilB, imipenem vs. exoU, exoS, nan1, and pilB, meropenem vs. exoU and pilB, gentamicin vs. exoU and pilB, tobramycin vs. exoU and pilB, amikacin vs. exoU and pilB, netilmicin vs. exoU and pilB, ciprofloxacin vs. exoU, exoS, and pilB, levofloxacin vs. lasB, exoU, and exoS, norfloxacin vs. exoU and exoS, lomefloxacin vs. exoU, and ofloxacin vs. exoS (P value < 0.05).
| Antibiotic Category/Antibiotic Agent | Antibiotic Resistance Rate; No. (%) | |
|---|---|---|
| Susceptible | Resistant a | |
| Penicillins | ||
| Piperacillin | 36 (42.9) | 48 (57.1) |
| β-lactam combination agents | ||
| Piperacillin-tazobactam | 45 (53.6) | 39 (46.4) |
| Ticarcillin-clavulanate | 5 (6) | 79 (94) |
| Cephems | ||
| Ceftazidime | 45 (53.5) | 39 (46.5) |
| Cefepime | 42 (50) | 42 (50) |
| Monobactams | ||
| Aztreonam | 48 (57.1) | 36 (42.9) |
| Carbapenems | ||
| Doripenem | 56 (66.7) | 28 (33.3) |
| Imipenem | 28 (33.4) | 56 (66.7) |
| Meropenem | 48 (57.1) | 36 (42.9) |
| Aminoglycosides | ||
| Gentamicin | 51 (60.7) | 33 (39.3) |
| Tobramycin | 52 (61.9) | 32 (38.1) |
| Amikacin | 43 (51.2) | 41 (48.8) |
| Netilmicin | 41 (48.8) | 43 (51.2) |
| Fluoroquinolones | ||
| Ciprofloxacin | 38 (45.2) | 46 (54.8) |
| Levofloxacin | 40 (47.6) | 44 (52.4) |
| Norfloxacin | 40 (47.6) | 44 (52.4) |
| Lomefloxacin | 28 (33.3) | 56 (66.7) |
| Ofloxacin | 20 (23.8) | 64 (76.2) |
aPseudomonas aeruginosa strains with intermediate sensitivity were considered as resistant isolates.
4.3. Virulence Genes Profile
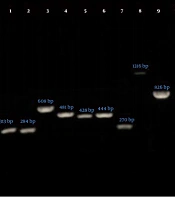
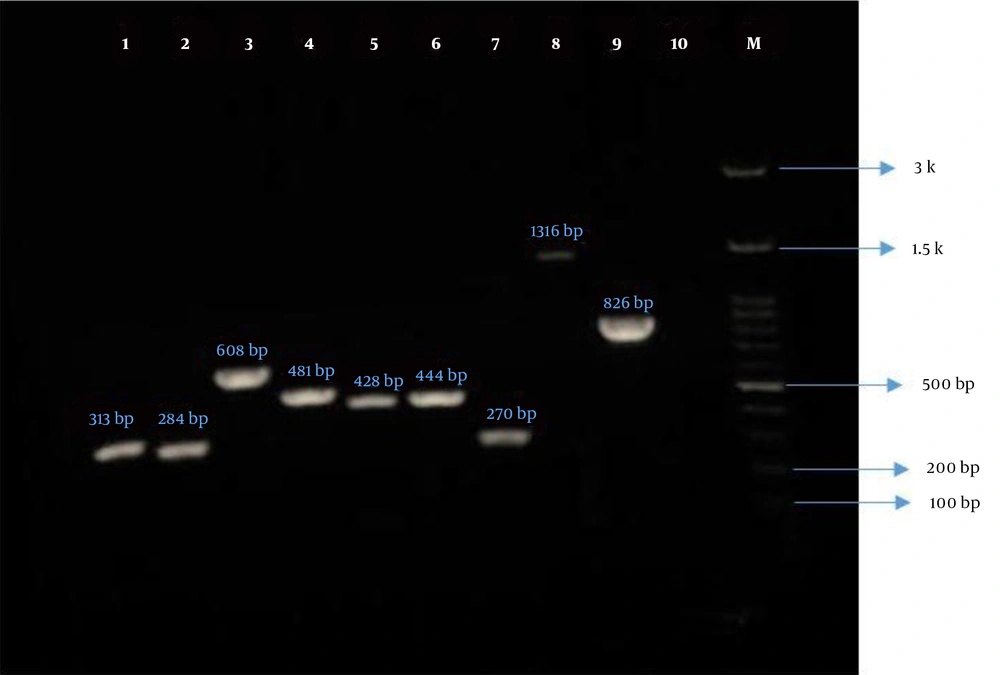
The frequency of virulence genes of P. aeruginosa was assessed using the PCR method and confirmed by sequencing (Figure 1). Nucleotide sequences were submitted to the GenBank database and the accession numbers were received (OK146924 to OK146931). The total prevalence of virulence genes of P. aeruginosa was as follows: (1) algD 84.5%, (2) lasB 86.9%, (3) plcH 86.9%, (4) plcN 86.9%, (5) exoU 56%, (6) exoS 51.2%, (7) toxA 81%, (8) nan1 13.1%, and (9) pilB 33.3%. Based on the findings of this study, there was no statistically significant association between the presence of virulence genes and sample type, except for the plcH and plcN genes (P = 0.02). Additionally, there was no significant difference in the prevalence of virulence genes between different genders and hospital wards (Table 3). Noteworthy, only one strain of P. aeruginosa isolated from the urine specimen of a (male) child hospitalized in the pediatric ward simultaneously contained all evaluated virulence genes (nine genes). As shown in Table 3, the highest prevalence rate of algD, lasB, plcH, plcN, exoU, exoS, toxA, nan1, and pilB genes was seen in urine specimens. Furthermore, isolates containing these genes were mostly recovered from patients hospitalized in the ICU and internal wards.
Gel electrophoresis of PCR products of P. aeruginosa virulence genes. Lane 1, algD (313 bp); Lane 2, lasB (284 bp); Lane 3, plcH (608 bp); Lane 4, plcN (481 bp); Lane 5, exoU (428 bp); Lane 6, exoS (444 bp); Lane 7, toxA (270 bp); Lane 8, nan1 (1316 bp); Lane 9, pilB (826 bp); Lane 10, negative control; and Lane M, ladder (100 bp).
| Gene | Gender (%) | P Value | Sample Source (%) | P Value | Ward (%) | P Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Urine | Blood | Wound | Sputum | ICU | Emergency | Internal | Neurology | Pediatric | ||||
| algD | 49.3 | 50.7 | 0.472 | 53.5 | 18.3 | 11.3 | 16.9 | 0.093 | 28.2 | 19.7 | 33.8 | 15.5 | 2.8 | 0.120 |
| lasB | 41.1 | 58.9 | 0.074 | 57.5 | 15.1 | 9.6 | 17.8 | 0.091 | 34.2 | 15.1 | 30.1 | 17.8 | 2.8 | 0.791 |
| plcH | 43.8 | 56.2 | 0.254 | 53.4 | 17.8 | 11 | 17.8 | 0.029 | 32.9 | 15.1 | 34.2 | 15.1 | 2.7 | 0.529 |
| plcN | 43.8 | 56.2 | 0.254 | 54.8 | 17.8 | 9.6 | 17.8 | 0.029 | 31.5 | 22 | 26 | 17.8 | 2.7 | 0.529 |
| exoU | 49 | 51 | 0.867 | 63.9 | 4.3 | 12.7 | 19.1 | 0.052 | 29.8 | 12.8 | 29.8 | 23.4 | 4.2 | 0.631 |
| exoS | 39.5 | 60.5 | 0.129 | 51.1 | 20.9 | 7 | 21 | 0.399 | 34.9 | 9.3 | 27.9 | 23.2 | 4.7 | 0.549 |
| toxA | 44.1 | 55.2 | 0.442 | 57.4 | 14.7 | 10.3 | 17.6 | 0.124 | 30.9 | 16.2 | 33.8 | 16.2 | 2.9 | 0.516 |
| nan1 | 54.5 | 45.5 | 0.254 | 45.4 | 27.3 | 18.2 | 9.1 | 0.415 | 27.3 | 18.2 | 36.3 | 9.1 | 9.1 | 0.362 |
| pilB | 53.5 | 46.5 | 0.440 | 67.9 | 17.8 | 10.7 | 3.6 | 0.107 | 28.6 | 21.5 | 39.3 | 7.1 | 3.5 | 0.230 |
5. Discussion
The emergence of bacterial antibiotic resistance is a growing threat to public health all over the world, especially in developing countries (14). According to predictions, antibiotic-resistant infections may lead to 10 million deaths per year by 2050 (14). Antibiotic resistance in P. aeruginosa is an important issue, particularly in patients with cystic fibrosis as well as hospitalized and immunocompromised patients (5). In 2017, the World Health Organization (WHO) listed carbapenem-resistant P. aeruginosa strains among important pathogens for which there is a need for new antibiotics (15). On the other hand, infections caused by MDR P. aeruginosa are also a global public health issue (16). In the present study, the frequency of MDR P. aeruginosa was 55.9%, which is close to the average prevalence reported from Iran (58%) (17). Carbapenems along with fluoroquinolones are two effective treatments against severe infections caused by MDR P. aeruginosa in hospital settings (18, 19).
In the current study, the prevalence of P. aeruginosa resistance to carbapenems was high (Table 2). Imipenem-resistant P. aeruginosa (66.7%) in this study was higher than those reported from Ahvaz (42.9%), Tabriz (49%), Urmia (30.8%), Zanjan (29.2%), Guilan (23.3%), Zahedan (17.2%), and Hamadan (7.5%), and lower than in Isfahan (76.1%) and Tehran (70.4%) (17). Additionally, the resistance rate to another carbapenem, i.e., meropenem, was high (42.9%) in Ardabil, which is comparable with that in Isfahan (93%), Tehran (78.8%), Ahvaz (44.1%), Urmia (39.4%), and Hamadan (13.2%) (17). Therefore, the use of carbapenems for the treatment of MDR P. aeruginosa infections is not recommended in Ardabil, except for doripenem (33.3%). Carbapenem-resistant P. aeruginosa strains indicated a high level of resistance to all β-lactam antibiotics, except for aztreonam (20). However, resistance to aztreonam was high in this study (42.9%). Similar results have been observed in other cities of Iran, including Ahvaz (91.3%), Tehran (83.7%), Isfahan (69%), Tabriz (60%), Urmia (56.3%), and Zanjan (37.5%), while the aztreonam-resistant rate reported from Zahedan was lower (14.7%) (17).
As seen in Table 2, resistance to other β-lactam antibiotics was high in Ardabil. Pseudomonas aeruginosa resistance to β-lactams can be attributed to β-lactamase enzymes, antibiotic efflux pumps, and reduced drug uptake (21). However, mechanisms of resistance to β-lactam antibiotics are not completely clear in local strains from Ardabil. As shown in Table 2, the resistance rate to fluoroquinolones was high in Ardabil city compared to other antibiotic classes. Fluoroquinolones, particularly ciprofloxacin, have remained as one of the most important antibiotics to treat a wide range of P. aeruginosa infections, including bacteremia, osteochondritis, ear and eye infections, external otitis, and chronic lung infections in cystic fibrosis patients (22). In this study, the prevalence of ciprofloxacin-resistant P. aeruginosa was lower in Ardabil (54.8%) than in Tehran (81.5%), Isfahan (78.7%), Guilan (66.3%), and Tabriz (65%), while it was higher than in Ahvaz (46.8%), Urmia (34.2%), Zanjan (32.5%), Hamadan (4.7%), and Zahedan (3.4%) (17).
Mutations in the ciprofloxacin target-encoding genes gyrAB and parCE and efflux pump overexpression are associated with resistance to fluoroquinolones (22). Amino acid alterations in the GyrA (Thr83Ile and Asp87Asn) and ParC (Ser87Leu and Ser87Trp) subunits of DNA gyrase and topoisomerase IV enzymes are involved in P. aeruginosa resistance to ciprofloxacin in Ardabil (19). Some aminoglycosides, such as amikacin and tobramycin, are commonly used for the treatment of pulmonary infections in patients with cystic fibrosis (21). Altogether, the aminoglycoside-resistant P. aeruginosa rate was found to be high in this study (Table 2). However, it seems that gentamicin and tobramycin are more effective than netilmicin and amikacin against P. aeruginosa infections in Ardabil. Aminoglycoside-modifying enzymes, rRNA methylases, and efflux pumps are involved in P. aeruginosa resistance to aminoglycosides (21). Overall, differences in P. aeruginosa drug resistance rates between this study and other studies in Iran can be attributed to self-medication, as well as inappropriate prescription and overuse of antibiotics.
Irrational use of antibiotics and the ensuing rise in the prevalence of antibiotic resistance, especially MDR, in clinical isolates of P. aeruginosa in Ardabil hospitals have a significant public health impact and may lead to increased hospitalization period medical costs, and mortality. On the other hand, it has been suggested that there is a relationship between drug resistance and virulence-associated genes in opportunistic bacteria such as drug-resistant P. aeruginosa (23). In the current study, a significant association was observed between resistance to some antibiotics and the prevalence of virulence genes in P. aeruginosa. These resistant and highly virulent bacteria can easily colonize in new environments or specific ecological niches with high antibiotic pressure, such as hospitals, and can cause diseases more efficiently (23). The current findings suggest that the prevalence of three virulence genes, i.e., lasB, plcH, and plcN (86.9%), in P. aeruginosa was higher than that of other virulence-associated genes in Ardabil. The most prevalent virulence genes of P. aeruginosa observed in other studies in Iran were as follows: (1) lasB gene (95.4%) (11); (2) exoS gene (92.9%) (12); (3) toxA gene (79.4%) (24); (4) lasB gene (92.9%) (25); (5) toxA gene (100%) (26); (6) lasB gene (100%) (27); and (7) lasB gene (100%) (28).
5.1. Conclusions
The current study revealed a high prevalence of resistance to all assessed antibiotics, except for doripenem, gentamicin, and tobramycin, in clinical isolates of P. aeruginosa. On the other hand, the number of MDR isolates was also alarmingly high. The highly virulent strains were prevalent in P. aeruginosa isolated from different specimens and hospital wards. This condition can be problematic for the efficient treatment of P. aeruginosa-associated infections in Ardabil hospitals. Hence, the continuous monitoring of P. aeruginosa isolates in terms of drug resistance trend, and virulence gene profile is needed.