Dear Editor,
Acquired hospital infection is the most important issue about the pathogenesis of Enterococcus faecalis, which causes a highly antibiotic-resistant disease (1). Due to the sharp increase in antibiotic resistance in E. faecalis, surveillance and supervision of this bacterium is very critical in the clinical setting. Accordingly, genotyping methods are very helpful in diagnosis and control of the infection. Also, the epidemiological surveillance of nosocomial infections is the main purpose of bacterial typing (2). In particular, pulsed-field gel electrophoresis (PFGE) is considered as a gold standard for the subtyping of enterococci (3). The PFGE method has been used to investigate the outbreak caused by enterococci along with other molecular typing methods, such as multiple-locus variable number tandem repeat analysis (MLVA), PCR ribotyping, restriction fragment length polymorphism (RFLP) and multi-locus sequence typing (MLST), but surely PFGE is the most preferable typing method (4).
It is also used as the most widely used typing method for the characterization of outbreaks due to vancomycin-resistant enterococci (VRE). Today, the PFGE method is the superior technique to track the origin of isolates due to the direct correlation with the genetic sequence of strains. In addition, it is able to nicely type bacterial isolates, and it has high repeatability of the results (5). The study and surveillance of E. faecalis strains are very necessary.
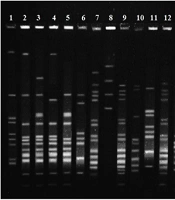
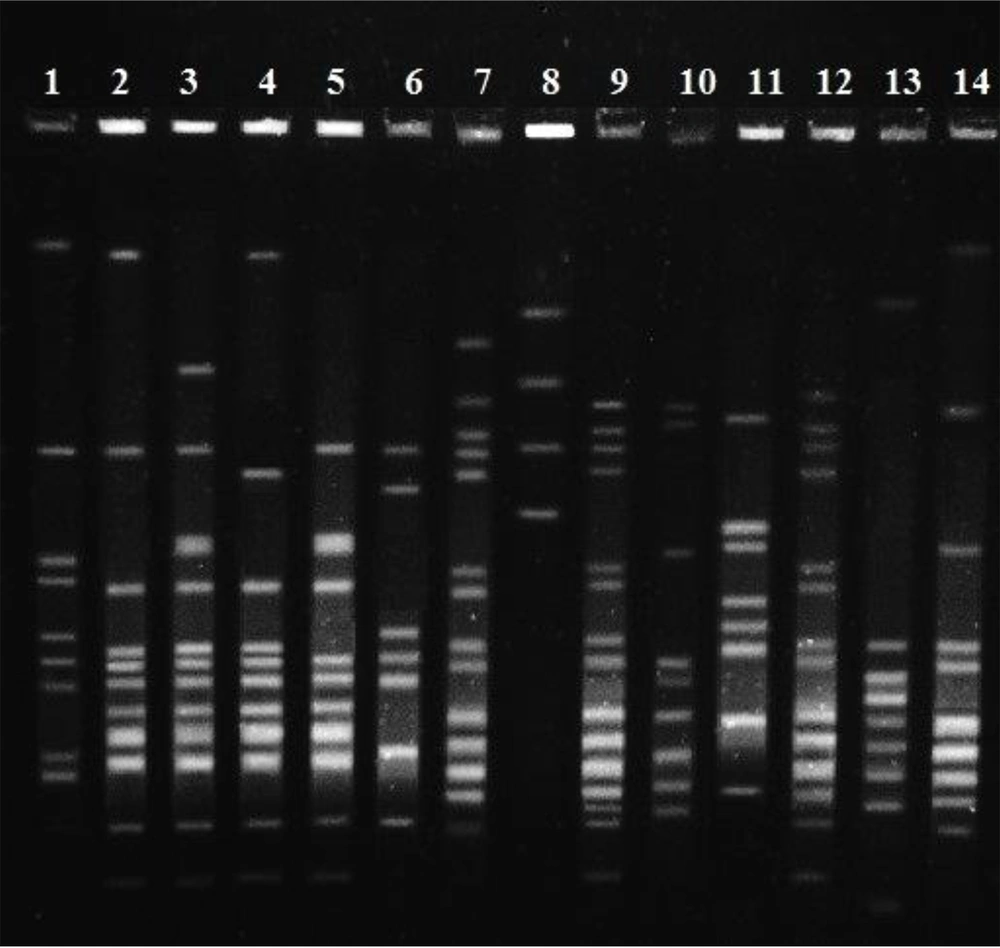
This study efforts to identify isolated haplotypes of Ilam, Iran by PFGE. The samples were from our previous study in which we studied the resistance of this bacterium to vancomycin (6). The main idea of the PFGE is separating a large fragment of the DNA, and the major part of PFGE is preparing agarose plugs. Each plug contains genomic DNA that is digested by Smal enzyme to obtain the appropriate fragment. Thus, plugs were digested by Smal enzyme for 4 hours at 25°C. In particular, contour-clamped homogeneous electric field (CHEF) electrophoresis is a technique of pulsed-field gel electrophoresis.
In order for staining, ethidium bromide was applied for 40 min, and then the gel was photographed. The results of the electrophoresis of each isolate were interpreted with cut off = 80%. The results of disk diffusion from our previous study (6) showed that 54% of the isolates were resistant to vancomycin. Ninety E. faecalis isolates that were sensitive and resistant to vancomycin were analyzed by the PFGE method. According to the cut off = 80, 26 different pulsotypes were observed. In detail, 17 pulsotypes had 2 isolates, and among 56 isolates, 9 different pulsotypes were seen, whereby two isolates could not be typed by the PFGE method. The results of our study demonstrated that 34 isolates belonged to the 17 pulsotypes, and all were sensitive to vancomycin. Also, there were 2 isolates that were non-typeable by PFGE and were sensitive to vancomycin. Ultimately, 8 vancomycin-sensitive isolates were located in different pulsotypes. In contrast, all vancomycin-resistant isolates are located at disparate pulsotypes (Figure 1).
The assessment by PFGE demonstrated that 48 out of 90 isolates had pulsotypes. It can indicate the different origins of E. faecalis clones. Overall, 26 various pulsotypes were observed, which indicated a poor correlation between isolates. The typing of E. faecalis has been done by other researchers using the PFGE method. In a research conducted by Bang et al. (7) to determine whether military working dogs (MWDs) are potential risk for human disease or not, distinguish patterns which resembled our results were reported.
Consist with the focus of this study, the gained information from isolation typing by the PFGE method is complementary to extensive epidemiological research, and it is valuable along with other genetic methods such as PCR at the different locations and times. In conclusion, our results showed that despite there was a VRE among isolates, but there were no clonal lineages in E. faecalis in Imam Khomeini Hospital. Also previous research that performed by MLST (6) confirmed the current results by PFGE.