1. Background
Infections caused by Acinetobacter baumannii are growing concerns in microbiology science and cause life-threatening diseases involving various organs (1, 2). Acinetobacter baumannii is rapidly developing resistance mechanisms to antibiotics. Extensive changes in resistance profile, especially against carbapenems, the medicine of choice to treat and control nosocomial infections with A. baumannii, have resulted in a high mortality rate and economic burden worldwide. Therefore, the multidrug-resistant (MDR) isolates of A. baumannii are a global problem in patients, particularly in intensive care units (ICUs) (1, 3).
Various mechanisms are employed to resist different antimicrobial agents, one of the most notable of which is integron, especially in gram-negative bacteria. The five main classes of integrons have been described based on the sequence identity of the int gene. Classes 1, 2, and 3 have an essential role in disseminating antimicrobial resistance genes, leading to the emergence of MDR phenotypes of A. baumannii. Integrons are mobile DNA elements that can integrate resistance gene cassettes and subsequently cause resistance phenotype in their bacterial host. An integron entails three genetic elements, namely integrase gene (intI), attachment site (attI), and promoter (Pc). The integrase gene is a tyrosine recombinase responsible for the site-specific recombination of mobile gene cassettes. Integrons are structurally composed of a 5' conserved segment (5'CS), a 3'conserved segment (3'CS), and an internal variable region consisting of one or more resistance genes cassettes captured by integron. Integrons are motionless but are contained in transposons and plasmids, allowing transfer through these mobile genetic elements (3-7). Integrons are the most prevalent genetic elements in the capture and accumulation of many antibiotic resistance genes in A. baumannii clinical isolates. As a result, studying integrons is a valuable method to investigate the molecular epidemiology of nosocomial outbreaks caused by this bacterium in the critical wards of hospitals, such as ICU (3-6).
Gene cassettes are mobile non-replicating elements consisting of an open reading frame and an attC site and are circular when not integrated into a cassette array. They often contain one or more antibiotic resistance genes and can be found either free or integrated at the attI site. Gene cassettes have an integrase-specific recombination site called the attC site. Recombination between the integron associated-intI site and attC sites leads to the insertion of the gene cassette downstream of a resident promoter mediated by the intI gene (7). As a result, gene cassettes containing antibiotic resistance genes are associated with MDR patterns in A. baumannii, resulting in outbreaks and therapeutic failure in healthcare settings (7). Therefore, assessing antimicrobial susceptibility profiles and detecting gene cassettes in the clinical isolates of A. baumannii in each geographical zone highlights the importance of epidemiological studies for the effective treatment and reasonable control of MDR and pandrug-resistant species of A. baumannii in hospital outbreaks (8, 9).
2. Objectives
There have been limited studies on the antibiotic resistance of A. baumannii due to integrons and gene cassettes in the southwestern region of Iran in recent years. The latest epidemiological survey in the southwest of Iran was conducted almost ten years ago, which determined the antimicrobial activity of conventional antibiotics against the isolates and the existence of integrons. With this background in mind, the present study aimed to evaluate antimicrobial susceptibility patterns, the presence of integrons, and associated gene cassettes among A. baumannii isolates obtained from hospitalized patients in the southwest of Iran.
3. Methods
3.1. Bacterial Isolates
All clinical strains examined in this study were isolated during July - September 2016 and were submitted to the Clinical Microbiology Laboratory of Nemazi Hospital and Prof. Alborzi Microbiology Research Center in Shiraz. The collected samples included sputum, blood, urine, throat swab, ulcer, endotracheal tube, lung biopsy, abdomen, axillary lymph node, as well as eye and nasal discharge. A total of 181 specimens were collected. Clinical samples of the patients were cultured on blood agar (Conda, Spain) and MacConkey agar (Conda, Spain) and were incubated for 16-18 h at 37°C (10). After the initial differential tests and, if suspected to be Acinetobacter spp, they were transferred to the sterile tubes containing tryptic soy broth medium (Conda, Spain). All bacteria were stored at -70°C. It should be noted that Acinetobacter spp. cannot be identified and confirmed at species level using conventional biochemical tests in clinical laboratories. To identify the isolates, we should consider bacterial-specific ribosomal gene replication.
However, morphological and biochemical tests were performed for the initial diagnosis of Acinetobacter. These tests included Gram staining, observing Gram-negative coccobacilli under a microscope, culture on an agar medium, observing small to medium convex, white or gray, and non-hemolytic colonies on triple sugar iron agar medium, and observing the growth pattern of ALK/ALK and H2S-negative bacteria. In addition, the bacterium was catalase-positive (catalase test reagent: Bahar afshan, Iran), oxidase-negative (oxidase test powder: Sigma, Germany), non-motile, indole-negative, MR (-) VP (-) in Voges–Proskauer test, nitrate-negative, citrate-positive, and produced acid in the oxidation-fermentation test (11). All the culture media were obtained from Conda, Spain.
3.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility patterns of the isolates were assessed using the Kirby-Bauer disk diffusion method on Mueller-Hinton agar as described by the Clinical and Laboratory Standards Institute (CLSI) (12, 13) guideline. All the isolates were sub-cultured from freezer stocks on sheep blood agar to prepare inoculum suspension. Following 18 - 24 h of incubation at 37°C on blood agar, the colonies were harvested to prepare a suspension. Bacterial isolates were suspended in sterile 0.85% saline to adjust turbidity at 0.5 McFarland standard equivalents to 1.5 × 108 CFU/mL. Escherichia coli (ATCC 25922) and A. baumannii (ATCC 19606) were used as control strains.
Ten antibiotic discs were placed on Mueller-Hinton agar (Conda, Spain) inoculated by bacterial suspension formerly. The plates were incubated for 24 h at 37°C. After incubation, the diameters of the inhibition zone were measured, and the isolates were classified as susceptible (S), intermediate (I), and resistant (R) based on the datasheet of the manufacturer. The antibiotic discs included trimethoprim-sulfamethoxazole (1.25/23.75 μg), amikacin (30 μg), gentamicin (10 μg), ampicillin-sulbactam (10/10 μg), pipracillin (100 μg), imipenem (10 μg), ceftazidime (30 μg), cefepime (30 μg), tetracyclin (30 μg), meropenem (10 μg), and polymyxin B (300 units). All discs were purchased from Mast Group Ltd, UK.
3.3. Polymerase Chain Reaction (PCR)
The isolates were identified as A. baumannii based on their molecular characteristics. Diagnosis of A. baumannii isolates at species level was confirmed by polymerase chain reaction (PCR) (Bio-Rad, T100 thermal cycler, USA) using the specific primers (Bioneer, South Korea) for A. baumannii OXA-51-like (F: 5′-TAA TGC TTT GAT CGG CCT TG-3′) and (R: CTTCGTGGATTCGACTTCAT) (350 bp) (14). Genomic DNA was extracted according to the method of Dashti et al. (15) with some modifications. The procedure was as follows: 2 or 3 colonies of an overnight culture of A. baumannii on brain heart infusion agar (Conda, Spain) were suspended in 200 µL of sterile distilled water and were boiled at 95°C for 10 min. The next step was removing cell debris by centrifugation for 10 min at 14000 rpm (Sigma, Germany). Afterward, 100 µL of supernatant was stored at -20°C for DNA amplification. The supernatant was analyzed for purity (carbohydrate and protein contamination), and A. baumannii genomic DNA concentration was assessed using the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, USA).
The PCR conditions were as follows: initial denaturation at 95°C for 5 min, 30 cycles of 95°C for 60 s, 50°C for 50 s, and 72°C for 50 s, followed by an elongation step at 72°C for 60 s. The total volume was 12.5 µL, and the PCR products were visualized by agarose gel electrophoresis (1.5% agarose gel) (Fanavaran Akhtarian, Iran). Sterile distilled water and the standard strain of A. baumannii (ATCC 19606) were used as negative and positive controls, respectively. The PCR products were electrophoresed on 2% agarose gel, containing DNA-safe stain (Pishgam, Iran) and visualized under UV light using a gel documentation system (Bio-Rad gel documentation system, USA).
3.4. Molecular Characterization of Class 1, 2, and 3 Integrons in Acinetobacter baumannii
PCR amplification of classes 1, 2, and 3 integrons was completed with the set of primers described by Goldstein et al. (16-18) with some modifications in the temperature settings with a total volume of 12.5 µL. A single PCR for detecting intI1 and duplex-PCR to identify intI2 and intI3 genes were carried out in the Master cycler gradient (BioRad, T100, USA) using the primers described previously. Primer sequences are shown in Table 1. Each reaction was completed in a final volume of 12.5 µL containing 6 µL of master mix (Amplicon, Denmark), 0.5 µL of each forward and reverse primers, 3 µL of template DNA, and 3 µL of distilled water. PCR was performed under the following conditions: initial denaturation at 94°C for 4 min (for intI1 gene) and 95°C for 5 min (for intI2 and intI3 genes) followed by denaturation at 94°C for 40 s (for intI1 gene) and 95°C for 1 min (for intI2 and intI3 genes), annealing at 58°C for 40 s (for intI1 gene) and 60°C for 50 s (for intI2 and intI3 genes), and extension at 72°C for 1 min. A final extension step was conducted at 72°C for 5 min. As mentioned previously (16-18), amplified products were visualized by gel electrophoresis.
| Genes | Sequence (5′→3′) | Product Size (bp) |
|---|---|---|
| Oxa51-like | 350 | |
| Forward | TAATGCTTTGATCGGCCTTG | |
| Reverse | CTTCGTGGATTCGACTTCAT | |
| intI1 | 280 | |
| Forward | CCTCCCGCACGATGATC | |
| Reverse | TCCACGCATCGTCAGGC | |
| intI2 | 233 | |
| Forward | TTATTGCTGGGATTAGGC | |
| Reverse | ACGGCTACCCTCTGTTATC | |
| intI3 | 600 | |
| Forward | AGTGGGTGGCGAATGAGTG | |
| Reverse | TGTTCTTGTATCGGCAGGTG |
Primers Applied for PCR Amplification
3.5. Detection of Class 1 Integron Gene Cassette Amplicons
We amplified variable regions of type 1 integron to detect the gene cassettes using primers described by Levesque and Roy (19).
3.6. Sequencing of Class 1 Integron Gene Cassette
The variable region of type 1 integron amplicons yielded from the previous step was sequenced in Macrogen (Korea), and nucleotide sequence alignment and comparisons were carried out using Basic Local Alignment Search Tool (BLAST) on the National Center for Biotechnology Information (NCBI) (https://www.blast.ncbi.nlm.nih.gov).
3.7. Statistical Analysis
Statistical analyses were performed by the student’s t-test, Fisher’s exact test, and chi-square test using the SPSS software version 18. P < 0.05 was considered statistically significant.
4. Results
A total of 174 isolates were included in the present study. The isolates were collected from 112 (64.3%) male and 62 (35.7%) female patients with a mean age of 51 ± 26 years. Most isolates were recovered from patients in the age range of 61 - 70 years. The isolates were mainly obtained from the ICU (50.9%, n: 87), followed by internal wards (27.01%, n: 47) and surgery wards (6.89%, n: 12). The most frequent specimens were respiratory secretion (27.5%, n: 48), endotracheal tubes (21.8%, n: 38), and blood (16.6%, n: 29).
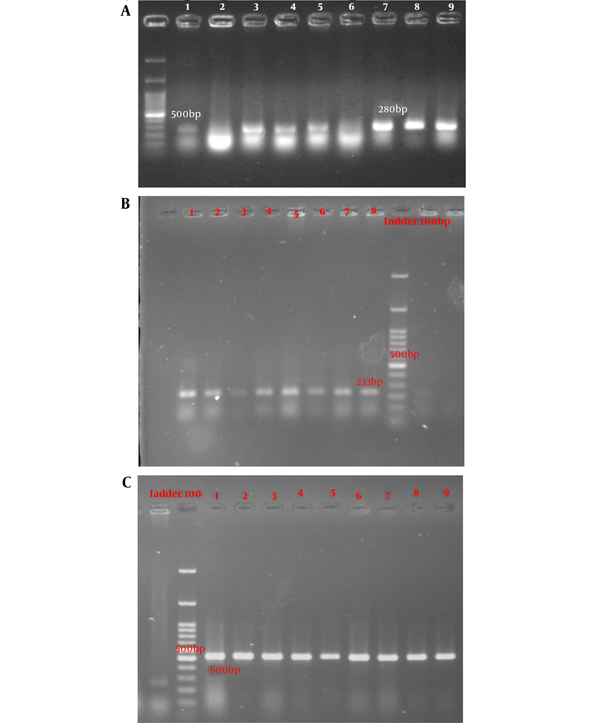
According to the PCR mapping results using the forward and reverse primers of the classes 1, 2, and 3 of integrons, 90.2% (n: 157), 72.4% (n: 126), and 12.1% (n: 21) of the isolates were positive for intI (Figure 1A), intII (Figure 1B), and intIII (Figure 1C) genes, respectively. We found that the age and gender of the patients were significantly correlated with the presence of the intII gene as this gene was merely detected in males (P = 0.001) and younger patients (P = 0.018). Moreover, a significant correlation was observed between the resistance of isolates to ampicillin-sulbactam and the presence of the intI gene (P = 0.003). Resistance to gentamycin and ceftazidime was significantly correlated with the presence of the intII gene (P = 0.05 and 0.02, respectively). In addition, the resistance of isolates to ceftazidime, tetracyclin, and cefepime had a significant correlation with the presence of the intIII gene (P = 0.02, 0.05, and 0.02, respectively).
Results of antimicrobial susceptibility testing in this study showed that polymixin B was the most effective antimicrobial agent against A. baumannii isolates. Furthermore, more than 90% of isolates (99.4% of isolates) were resistant to imipenem and it was the highest level of resistance to a antibiotic between all the antibiotics were tested in this research. The relationship between integrons and antibiotic resistance in Acinetobacter isolates is shown in Table 2. Statistical analysis revealed the gender of patients to be significantly correlated with the antibiotic susceptibility patterns of co-trimoxazole, ceftazidime, and amikacin. In this regard, resistance to the three mentioned antibiotics was significantly higher in males (P = 0.027, 0.01, and 0.01, respectively). Moreover, the analysis of variance demonstrated a significant correlation between age and susceptibility to tetracycline (P = 0.01).
| No. | Antibiotics Resistant Pattern (No. of Antibiotics) | No. of Isolates | Integron Class 1 | Integron Class 2 | Integron Class 3 |
|---|---|---|---|---|---|
| 1 | CPM/T/CAZ/PB/GM/PRL/TS/AK/MEM/IMI (10) | 1 | 1 | 0 | 0 |
| 2 | CPM/T/SAM/CAZ/GM/PRL/TS/AK/MEM/IMI (10) | 56 | 51 | 38 | 8 |
| 3 | CPM/SAM/CAZ/GM/PRL/TS/AK/MEM/IMI (9) | 54 | 48 | 46 | 7 |
| 4 | CPM/T/CAZ/GM/PRL/TS/AK/MEM/IMI (9) | 17 | 16 | 10 | 1 |
| 5 | T/SAM/CAZ/GM/PRL/TS/AK/MEM/IMI (9) | 6 | 6 | 4 | 0 |
| 6 | CPM/T/SAM/CAZ/GM/PRL/AK/MEM/IMI (9) | 8 | 7 | 4 | 1 |
| 7 | CPM/T/SAM/GM/PRL/TS/AK/MEM/IMI (9) | 4 | 4 | 3 | 0 |
| 8 | CPM/T/SAM/CAZ/GM/AK/MEM/IMI (8) | 1 | 1 | 0 | 0 |
| 9 | CPM/T/CAZ/GM/PRL/AK/MEM/IMI (8) | 3 | 2 | 1 | 9 |
| 10 | T/CAZ/GM/PRL/TS/AK/MEM/IMI (8) | 5 | 5 | 0 | 0 |
| 11 | CPM/CAZ/GM/PRL/TS/AK/MEM/IMI (8) | 4 | 3 | 2 | 1 |
| 12 | CPM/T/SAM/CAZ/PRL/TS/MEM/IMI (8) | 1 | 1 | 0 | 0 |
| 13 | CPM/T/CAZ/GM/PRL/AK/MEM/IMI (8) | 2 | 2 | 0 | 0 |
| 14 | CPM/SAM/CAZ/GM/PRL/AK/MEM/IMI (8) | 1 | 1 | 0 | 1 |
| 15 | CPM/T/CAZ/GM/PRL/TS/MEM/IMI (8) | 1 | 1 | 0 | 0 |
| 16 | T/CAZ/GM/PRL/AK/MEM/IMI (7) | 4 | 4 | 0 | 1 |
| 17 | T/GM/PRL/TS/AK/MEM/IMI (7) | 1 | 1 | 1 | 1 |
| 18 | CPM/SAM/CAZ/GM/PRL/TS/IMI (7) | 1 | 1 | 1 | 0 |
| 19 | CAZ/PRL/TS/AK/MEM/IMI (6) | 1 | 1 | 1 | 0 |
| 20 | T/SAM/CAZ/GM/PRL/TS (6) | 1 | 1 | 1 | 0 |
| 21 | CAZ/TS/MEM/IMI (4) | 1 | 0 | 0 | 0 |
| 22 | T/GM/PRL/AK/MRM (4) | 1 | 1 | 1 | 0 |
MDR Phenotype of Acinetobacter baumannii Clinical Isolates for intI, II, and III
All isolates obtained from patients in this study were considered MDR based on the definition given by Magiorakos et al. The MDR phenotypes of the clinical isolates of classes 1, 2, and 3 integrons are shown in Table 3. Amplification of the variable region of integron class 1 produced seven different sizes of gene cassettes which contributed to integron class 1 with the size range of 500 - 1500 bp. Several amplicons with different lengths were selected and analyzed by sequencing, and the data were compared in GenBank. The results demonstrated that three separate isolates with the size of 1500 bp carried two distinct types of class 1 integron gene cassettes, including aadA2 and dfrA12. The related data are shown in Table 4. In addition, our results indicated that isolates with gene cassettes of the same size almost presented similar antimicrobial resistance patterns (Appendix 1).
| Class of Antimicrobial Agent | Resistant Isolates (%) | Presence of Integron 1 (P-Value) | Presence of Integron 2 (P-Value) | Presence of Integron 3 (P-Value) |
|---|---|---|---|---|
| Cephalosporins | ||||
| Cefepime | 88.5 | 0.58 | 0.06 | 0.02 |
| Ceftazidime | 95.4 | 0.56 | 0.02 | 0.02 |
| Tetracyclines | ||||
| Tetracycline | 63.2 | 2.2 | 8.7 | 0.05 |
| DHFR inhibitor/sulfonamide | ||||
| Trimethoprim/sulfamethoxazole (Co-trimoxazole) | 90.8 | 0.14 | 3.5 | 1.1 |
| Penicillins/beta-lactamase inhibitor | ||||
| Ampicillin/sulbactam | 76.4 | 0.003 | 1.4 | 0.71 |
| Polymyxins | ||||
| Polymyxin B | 0.57 | 0.1 | 2.6 | 0.12 |
| Beta-lactams | ||||
| Imipenem | 99.4 | 0.1 | 0.38 | 0.12 |
| Meropenem | 97.7 | 0.41 | 1.5 | 0.5 |
| Broad-spectrum beta-lactams | ||||
| Piperacillin | 98.2 | 1.92 | 2.3 | 0.37 |
| Aminoglycoside | ||||
| Gentamicin | 98.2 | 1.92 | 0.05 | 0.37 |
| Amikacin | 95.4 | 0.07 | 0.41 | 0.89 |
Association Between the Existence of Integrons and Antibiotic Resistance in 174 Clinical Isolates of Acinetobacter baumannii
| Pattern of Gene Cassettes Associated with intI (bp) | Number of Isolates | Gene Cassette |
|---|---|---|
| 500 | 8 | aadA1 |
| 700 | 17 | dfrA5, dfrA25 |
| 750 | 1 | aadB |
| 1000 | 6 | aadA1, aadA2 |
| 1200 | 16 | blaCARB-2 |
| 1400 | 13 | aadB-catB3 |
| 1500 | 3 | dfrA1–aadA1a |
Size of Amplicons and Gene Cassettes Associated with intI
5. Discussion
Bacterial infections are one of the leading causes of death worldwide. Lack of sensitivity to antibiotics can increase the risk of surgery and fundamental human health challenges. The most important genus of MDR bacteria that have recently intelligently escaped antimicrobial therapy and have spread as a nosocomial pathogen worldwide is called ESKAPE. One of the most important strains of this group is A. baumannii. Infections caused by this bacterium are significantly increasing in hospitals worldwide (20).
The clinical importance of A. baumannii arises from the rapid global emergence of MDR strains, resulting in a high mortality rate and difficulties for the health organization. Regarding the mentioned facts, epidemiological findings could help recognize the susceptibility profiles of A. baumannii, causing infections in different areas (21). Integrons have now been shown to be the primary carriers of multiple antibiotic resistance as mobile genetic elements in gram-negative bacteria and, less importantly, in gram-positive bacteria (22). The main purpose of the current study was to determine the frequency of integrons and gene cassettes related to antibiotic resistance in A. baumannii isolates from hospitalized patients and outpatients of Nemazi Hospital, Shiraz, and Prof. Alborzi Microbiology Research Center in 2016. The obtained results might be used to plan for preventive measures and prevent the spread of resistant strains.
As mentioned before, A. baumannii is currently an important cause of nosocomial infections. Consequently, samples were also collected from hospitalized patients in the present study. Our findings showed that half of the A. baumannii isolates (50.9%) were obtained from patients hospitalized in the ICU. This result is consistent with those of previous studies about the role of A. baumannii in ICU infections (23, 24). According to the literature, there is a direct relationship between the length of hospital stay and the likelihood of infection with A. baumannii (25, 26). It can be argued that the reason for the high prevalence of disease caused by A. baumannii in the ICUs is the long duration of hospitalization in these wards, making the patient more prone to nosocomial infections.
The antibiogram of A. baumannii isolates in the present study indicated the high resistance of these samples compared to other similar studies. We found more than 90% resistance against seven of the eleven investigated antibiotics, and the only antibiotic with less than 50% resistance was polymyxin B. One of the significant data obtained in this study was the high percentage of MDR phenotypes along with the high prevalence of integrons in the evaluated clinical isolates, representing the importance of integrons in disseminating antibiotic resistance genes in the environment (21, 27-29).
In this study, screening for MDR phenotypes among A. baumannii isolates showed an alarming elevating trend of resistance to multiple antibiotics. In this regard, Rolain et al., Cicek et al., and Moradi et al. have also demonstrated the high frequency of MDR phenotype in A. baumannii isolates in Qatar, Turkey, and Iran, respectively. One of the remarkable results of the present study compared to similar investigations was that 100% of the studied isolates had MDR phenotypes. It seems that the emergence of MDR strains, which is likely due to the improper use of antimicrobial agents, limits therapeutic protocols (30-32). In line with our study, several MDR isolates of A. baumannii have been reported from the hospitals of the United Arab Emirates, Bahrain, Saudi Arabia, Palestine, and Lebanon (33, 34).
As mentioned in the present study, 100% of the isolates had multiple antibiotic resistances. The MDR frequency among tested isolates is similar to that stated previously in Poland by Koczura et al. (100%) (35), in Greece by Kraniotoki et al. (100%) (36), and in China by Zhao et al. (93 %) and Huang et al. (37, 38). However, this is considerably higher than those reported in other recent reports from Thailand (21.1%) by Aimsaad et al. (39) and from China by Zheng et al. (61.3%) (40). This aspect should be considered in treating the relevant infections to prevent the inappropriate use of broad-spectrum antibiotics, which could cause more implications during the course of the disease.
The present study investigated the resistance of bacterial isolates to six different classes of antibiotics and co-trimoxazole (trimethoprim and sulfamethoxazole). The results showed 90% resistance to six antibiotics, namely piperacillin, amikacin, gentamicin, ceftazidime, imipenem, and meropenem, in four separate classes of antibiotics and co-trimoxazole. Therefore, the isolates could be considered extensively drug-resistant (XDR). A bacterial isolate resistant to carbapenems, in addition to three antibiotics from three separate classes, is called an XDR isolate (13). It should be noted that the epidemiological importance of XDR bacteria is not only because of their resistance to several antimicrobial agents but also because of their unique ability to become resistant to all or most of the antimicrobial agents (33).
This issue should be considered that resistance to carbapenems, including imipenem and meropenem, has dramatically increased compared to a previous study carried out in Shiraz by Japoni-Nejad et al. (41). Consistent with our results, carbapenem-resistant A. baumannii isolates were detected in a recent survey in Shiraz that investigated metallo-beta-lactamase enzymes production (42). Our findings showed significantly higher resistance rates to cephalosporins and aminoglycosides than previous reports. Therefore, the prescription of these antimicrobial groups should be reviewed to manage nosocomial infections. The results showed that polymixins (e.g., polymixin B) are the most effective antimicrobial agents, in agreement with previous reports (Velkov et al. and Genteluci et al.). Although the efficacy of polymixins has been declared in the literature, the prescription of these medications is confined because of their neurotoxic or nephrotoxic side effects (43, 44).
Moreover, in the present study, the highest antibiotic susceptibility after polymyxin was observed for tetracycline (36.8%). According to the studies by Lee et al. in Taiwan and Ni et al. in China, A. baumannii isolates are reasonably susceptible to the antibiotic tigecycline, a tetracycline derivative (45, 46). Tetracycline has historically been introduced much earlier than some other antibiotics, such as imipenem. However, resistance to older antibiotics is less common among bacterial isolates due to the overuse of imipenem and other beta-lactamase-resistant beta-lactams for MDR. Acquisition of foreign genetic elements, such as integrin leads to the emergence of resistant phenotypes. The spread of these mobile elements among bacterial species causes resistance in healthcare settings. We found a remarkable rise in the presence of class 1 and 2 integrons compared to the previous study in the same area by Japoni-Nejad et al. (41). This trend is acceptable considering the changes in the resistance patterns of isolates compared to the previous survey. Furthermore, our results are consistent with those of other investigations by Xu et al. in China and Japoni-Nejad et al. in Iran, representing a higher prevalence of integron class 1 than class 2 and class 3 (41, 47).
Statistical analysis showed a significant relationship between the presence of integron and resistance to antimicrobial agents, including ampicillin-sulbactam, cephalosporins, gentamycin, and tetracyclin. It should be noted that mechanisms other than integron acquisition are also involved in antibiotic resistance (48, 49). According to the results of the present study, 157 out of 174 bacterial isolates (90.2%) had class 1 integron. In similar studies performed by Xu et al. and Taherikalani et al. in China and Iran, 53 and 58% of the isolates contained class 1 integron, respectively (47, 50). The prevalence of class 1 integron in the current research is in accordance with the rate found by Peymani et al. in Iran (92.5%) (51).
The presence of class I integron in the present study was significantly correlated with ampicillin-sulbactam resistance. As a result, the augmentation in resistance to this class of antibiotics is consistent with the rise in the presence of class I integron observed in comparison with the previous study in the same area (41). Whereas none of the isolates in the previous studies harbored class 3 integron (52), 12.1% of our isolates contained this class of integron genes. As we found a significant correlation, the existence of the class 3 integron gene could be correlated with the increased resistance of studies isolates to cephalosporins. Indeed, these groups of antimicrobial agents should be prescribed more cautiously considering the global resistant dissemination (53). In addition, in a previous study in Iran by Japoni-Nejad, resistance to aminoglycosides was reported. Acquiring the class 3 integron gene may result in activated efflux pumps and resistance to tetracycline (54).
The present study is also the first investigation of the gene cassettes among the isolates of A. baumannii in Shiraz. Seven different sizes of integron class 1 gene cassettes were detected by PCR. The sequencing method for identifying the types of gene cassettes indicated two different types of class I integron gene cassettes, namely aadA2 and dfrA12, both of which had previously been reported. The dfrA12 is related to the expression of the dihydrofolate reductase gene, which is contributed to resistance to trimethoprim. In this study, 90.8% of the isolates were resistant to co-trimoxazole (trimethoprim-sulfamethoxazole). These gene cassettes have previously been reported in Iran and India by Japoni-Nejad et al. and Girija et al., respectively (41, 55).
The aadA2 is related to the expression of the aminoglycoside adenylyltransferase gene responsible for resistance to aminoglycoside antibiotics, such as amikacin and gentamycin. Our results revealed that 98.2 and 95.4% of the isolates were resistant to gentamycin and amikacin, respectively. This gene cassette has been reported in other regions in Iran (8, 41, 56), Turkey (31), and Australia (31, 57). Considering our results, resistant isolates to trimethoprim-sulfamethoxazole contained the dfrA12 gene cassettes. Furthermore, the presence of aadA2 gene cassettes is accompanied by resistance to gentamycin and amikacin. Consequently, it is worth noting to declare a significant relationship between the presence of gene cassette and reduced susceptibility to antibiotics. A small number of arrays related to class I integron gene cassettes were identified in the present study. However, sequencing the components specified by PCR was one of the advantages of the current research that has not been considered in many similar domestic studies, especially in southwestern Iran. We examined the resistance phenotype of A. baumannii isolates and observed 22 different phenotypes. In addition, class 1, 2, and 3 integrons were identified separately, and cassettes related to class 1 were assessed. In this regard, the present study is unprecedented in the southwestern region of Iran.
5.1. Conclusions
The results of the current investigation indicated a high prevalence of class 1 and 2 integrons in A. baumannii isolates and a significant prevalence of class 3 integrons compared to similar studies. This finding, along with the high antibiotic resistance of the studied isolates as 100% of the isolates were MDR, clearly indicates the importance of integrons in the spread of antibiotic resistance genes among bacteria. Sequencing results confirmed the existence of two cassette arrays of aadA2 and dfrA12. These arrays encode the enzymes resistant to aminoglycosides and trimethoprim. Resistant to most of the antibiotics evaluated in the present study is highly prevalent. Therefore, the wide administration of carbapenems (e.g., imipenem and meropenem) and cefepime in hospitals should be limited. Continuous monitoring and characterization of integrons and their associated gene cassettes could help control the rate of antibiotic resistance by planning preventive measures to hinder the spread of resistant strains.