1. Background
The most common bacteria causing urinary tract infections (UTI) are Escherichia coli and Klebsiella pneumoniae (1). Escherichia coli is responsible for more than 85% of all UTIs in hospital and community settings (2). Based on statistics, 150 million people suffer from UTIs each year worldwide. Urinary tract infections occur about 30 times more frequently in females than males, and nearly 60% of them are infected with UTIs at least once in their lifetime (3). Indiscriminate use of antibiotics is a contributing factor that often leads to multi-drug-resistant strains. The remarkable ability of E. coli and K. pneumoniae to obtain plasmids, integrons, or transposons with clusters of resistant genes can result in multidrug resistance (MDR). The most severe medical concern is the control of MDR because the current therapeutic choices are limited. The resistance to carbapenems increased sharply in the past decade (4, 5).
The high level of carbapenem-insensitive Gram-negative bacteria has become a concern worldwide (6). Carbapenem resistance can be attributed to one of the mechanisms causing resistance in bacteria, such as the inactivation of enzymes, overproduction of efflux pumps, and modification of target sites. Nevertheless, among the reported mechanisms causing carbapenem resistance, the most important mechanisms are carbapenem-hydrolyzing-β-lactamases associated with the metallo-β-lactamases (Ambler class B) and oxacillinases (Ambler class D). The ability to hydrolyze oxacillin can be an effective reaction (7, 8). The appearance of carbapenem-hydrolyzing class D β-lactamases depends on antimicrobial chemotherapy, particularly carbapenems. Major subgroups of acquisitive carbapenem-hydrolyzing class D β-lactamases include OXA-23-like, OXA-40-like, OXA-51-like, and OXA-58-like β-lactamases (9). The OXA-51-like lactamase has been discovered worldwide, e.g., in Argentina, Austria, England, France, Greece, Kuwait, Romania, Spain, Scotland, and Turkey (10).
El-Badawy et al. studied OXA-23, and OXA-51 β-lactamases in carbapenem-resistant K. pneumoniae isolates in Egypt and found that 5.3% and 10.5% of the isolates could produce OXA-23 and OXA-51 β-lactamases, respectively (11). In Cetinkol et al.’s study, OXA-23-like, OXA-40-like, OXA-51-like, and OXA-58-like β-lactamases were not observed in carbapenem-resistant K. pneumoniae isolates (12). Manohar et al. studied carbapenem resistance genes in Gram-negative bacteria in India and found the uncommon attendance of OXA-23 in E. coli (n = 4) and OXA-23 and OXA-51 in K. pneumoniae (13).
2. Objectives
This research examined the antimicrobial susceptibility patterns and OXA-23, OXA-24, OXA-40, OXA-51, and OXA-58 genes in clinical strains of E. coli and K. pneumoniae.
3. Methods
3.1. Isolates and Antimicrobial Susceptibility Tests
We collected 500 urine isolates from patients with UTI indications hospitalized at a public hospital in Tehran from 2019 to 2020. The reconfirmation of the isolates was performed by standard biochemical methods. The whole isolates were maintained at -80°C in trypticase soy broth with 15% glycerol for further processing. The minimum inhibitory concentration (MIC) was determined by the E-test method (Liofilchem® MIC Test Strips). The MIC was performed for ceftazidime (CAZ), ceftriaxone (CRO), meropenem (MEM), ciprofloxacin (CIP), piperacillin/tazobactam (TZP), gentamicin (GEN), amoxicillin-clavulanate (AMC), and trimethoprim/sulfamethoxazole (SXT). The data were interpreted as resistant, intermediate, and susceptible, conforming to the M100-CLSI 2021 instructions (14).
3.2. Confirmation of Carbapenemase Activity
The isolates were tested for carbapenemase activity with Carba-NP according to the CLSI guidelines and manufacturer instructions. The color change of the trial vial to perfect yellow or red-yellow confirmed carbapenem-resistant isolates, while the control vial stayed ruddy (15).
3.3. Recognition of OXA Group Genes
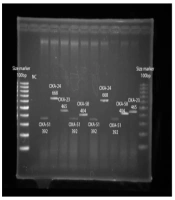
The genomic DNA of isolates was extracted by a High Pure PCR Template Preparation kit (Roche, Germany). Recognition of OXA group genes was done by formerly defined particular primer sets (Metabion, Germany) shown in Table 1. According to the manufacturer’s guidelines, the DNA amplification by PCR was done using a Peqlab PCR thermal cycler and PCR Master Mix (Ampliqon Inc., Denmark). The experiment was done in a final volume of 25 µL. The reaction mix contained 12 µL of Master Mix Red (Ampliqon Inc., Denmark) and 1 µL of template DNA. Forward and reverse primers were adjoined according to Table 1. The total volume of the reaction mixture was adjusted to 25 µL. Primary denaturation was regulated at 95°C for 5 min, followed by 30 cycles of 94°C for 25 s, the ideal annealing temperature per gene for 40 s, and 72°C for 50 s. Afterward, the final extension was done at 72°C for 5 min. Amplicons of PCR were separated by 1.5% w/v agarose gels (16).
| Studied Genes | Annealing Temperature °C | Amplicon Size | Reference |
|---|---|---|---|
| Oxa-51-like | 53 | 392 | (16) |
| F: TAATGCTTTGATCGGCCTTG | |||
| R: TGGATTGCACTTCATCTTGG | |||
| Oxa-23-like | 53 | 465 | (16) |
| F: GATCGGATTGGAGAACCAGA | |||
| R: ATTTCTGACCGCATTTCCAT | |||
| Oxa-24-like | 53 | 668 | (16) |
| F: GGTTAGTTGGCCCCCTTAAA | |||
| R: AGTTGAGCGAAAAGGGGATT | |||
| Oxa-58-like | 53 | 404 | (16) |
| F: AAGTATTGGGGCTTGTGCTG | |||
| R: CCCCTCTGCGCTCTACATAC |
Primer Sequences for OXA Group Genes
3.4.Multi Locus Variable-Number Tandem Repeat Analysis Technique in Escherichia coli
The overnight cultures of the isolates were applied for preparing total genomic DNA utilizing High Pure PCR Template Preparation kits (Roche, Germany). The multilocus variable number of tandem repeat analysis was performed utilizing seven loci (CVN001, CVN002, CVN003, CVN004, CVN007, CVN014, and CVN015) as previously explained by Lindstedt et al. (17). The previously published primers (17) were used for multi locus variable-number tandem repeat (VNTR) analysis (MLVA) (Table 2). The loci were multiplied by PCR and assessed with 3% agarose.
| Studied Loci | Annealing Temperature (°C) | Reference |
|---|---|---|
| CVN001 | 50 | (17, 18) |
| F: AACCGGCTGGGGCGAATCC | ||
| R: GGCGGCGGTGTCAGCAAATC | ||
| CVN002 | 50 | (17, 18) |
| F: AACCGTTATGAARGRAAGTCCT | ||
| R: TCGCCCAGTAAGTATGAAATC | ||
| CVN003 | 50 | (17, 18) |
| F: AAAAATCCGGATGAGWTGGTC | ||
| R: TTGCGTTGTCAGTAATTTGTTCAG | ||
| CVN004 | 50 | (17, 18) |
| F: MGCTGCGGCRCTGAAGAAGA | ||
| R: CCCGGCAGGCGAAGCATTGT | ||
| CVN007 | 50 | (17, 18) |
| F: ACCGTGGCTCCAGYTGATTTC | ||
| R: ACCAGTGTTGCGCCCAGTGTC | ||
| CVN014 | 50 | (17, 18) |
| F: TCCCCGCAATCAGCAAMACAAAGA | ||
| R: GCAGCRGGGACAACGGAAGC | ||
| CVN015 | 50 | (17, 18) |
| F: TAGGCATAGCGCACAGACAGATAA | ||
| R: GTACCGCCGAACTTCAACACTC | ||
| VNTR10 | 58 | (19, 20) |
| F: AGCGCGCAGACGATGAGCAG | ||
| R: AGCCCCGCAGTGGGGTTACT | ||
| VNTR27 | 58 | (19, 20) |
| F: CAGCGTCAGCGCCAGACCAA | ||
| R: CCATGGCCGGCCTGTGGTTT | ||
| VNTR45 | 58 | (19, 20) |
| F: CGCTGACACATTGACGAAAACAGAGA | ||
| R: ATGAATATTGCCCAGTTTCTGGAACAA | ||
| VNTR51 | 58 | (19, 20) |
| F: CCGCCGCGCCATCGTTAGAT | ||
| R: TCAACGCGCCCAGCTGAACC | ||
| VNTR52 | 58 | (19, 20) |
| F: TTTGGCGGCAGCGGTTTCCC | ||
| R: GCCAGAAAAAGGCGCGCAGC | ||
| VNTR53 | 58 | (19, 20) |
| F: CGCAGAAGAAAGCGGAAG | ||
| R: TGTTTTAGGCGCATTCTTACC | ||
| VNTR58 | 58 | (19, 20) |
| F: CTATCTGGCGAACCAGACG | ||
| R: ATTATGACGGGCGATATAATAGGC | ||
| VNTR60 | 58 | (19, 20) |
| F: CGGTACGAATCTGTTGGATTAAG | ||
| R: GGCCTTCTTCCGGGTCTAT |
Primer Sequences for MLVA Technique for Escherichia coli and Klebsiella pneumoniae
The numbers of VNTR repeats per locus were assessed by the following equation (17): (NPS - OF)/RL, wherever PS = product size, OF = offset region (region of sequence not containing repeat), and RL = length of one repeat unit. CVN001: ((PS) - 250)/39, CVN002: ((PS) - 272)/18, CVN003: ((PS) - 404)/15, CVN004: ((PS) - 231)/15, CVN007: ((PS) - 314)/18, CVN014: ((PS) - 111)/6, and CVN015: ((PS) - 189)/6 (17, 21). The numbers of VNTR repeats were rounded to the closest whole repeat.
3.5. Multi Locus Variable-Number Tandem Repeat Analysis Technique in Klebsiella pneumoniae
Klebsiella pneumoniae MLVA was performed using eight tandem sequence repeats, including VNTR10, VNTR27, VNTR45, VNTR51, VNTR52, VNTR53, VNTR58, and VNTR60, as described by Lindstedt et al. (22). Previously published primers were used for MLVA analysis (Table 2). The loci were multiplied by PCR and assessed with 3% agarose. The VNTR repeat numbers for each locus were calculated (13): (NPS - OF)/RL. These allele definitions were applied to analyze the clinical isolates using the MLVA plugin of the BioNumerics program (Version 6.6, Applied Maths, Sint-Martens-Latem, Belgium) (21).
3.6. Data Analysis Method
We utilized SPSS v.22 (SPSS Inc., Chicago, IL, USA) for statistical analysis. A P value < 0.05 was assumed statistically significant.
4. Results
We collected 500 urinary bacterial isolates of E. coli and K. pneumoniae. Women had more urinary tract infections (190 cases of K. pneumoniae and 150 cases of E. coli) than men (80 cases of K. pneumoniae and 80 cases of E. coli). The UTIs with E. coli were most prevalent in 70 - 79-year patients, while those with K. pneumoniae were most prevalent in 51 - 59-year patients.
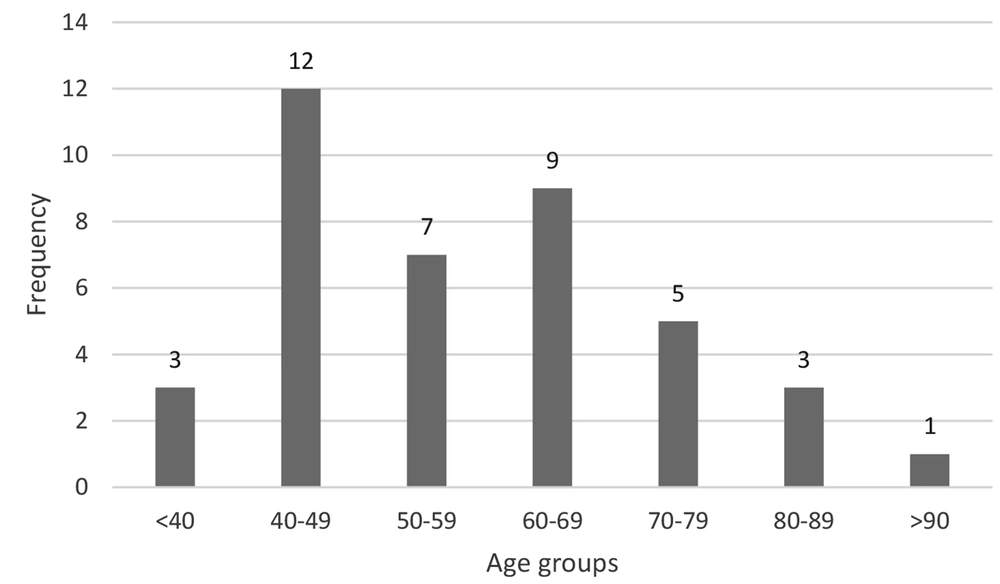
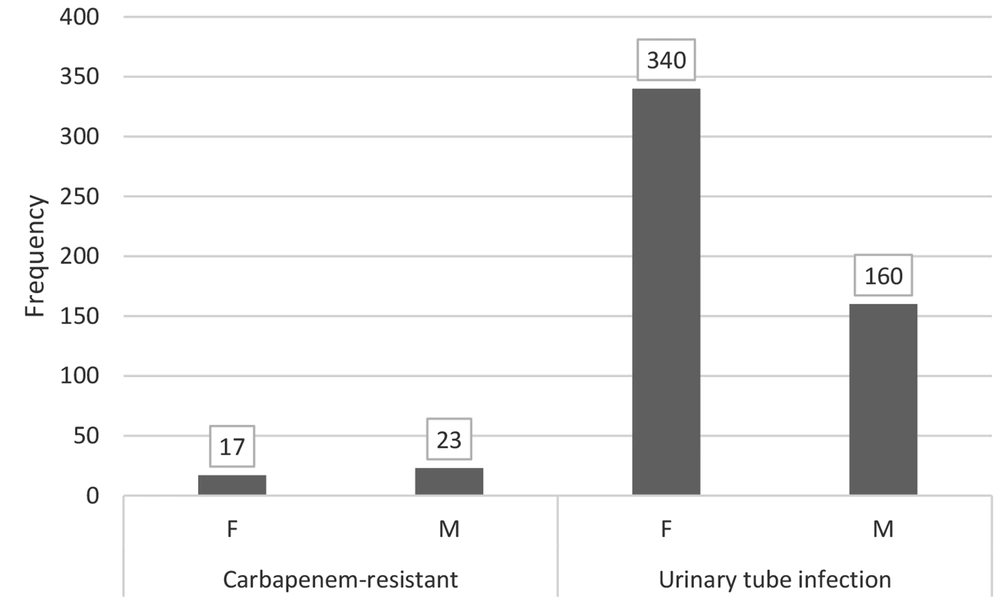
Out of 500 isolates, 40 (8%) were carbapenem-resistant, including 13 E. coli and 27 K. pneumoniae. Forty carbapenem-resistant isolates were used for the present study. The most common UTIs were observed in the 40 - 49-year age group (Figure 1). Interestingly, men had more UTIs (23 cases) than women (17 cases) (Figure 2).
4.1. Antimicrobial Susceptibility Testing
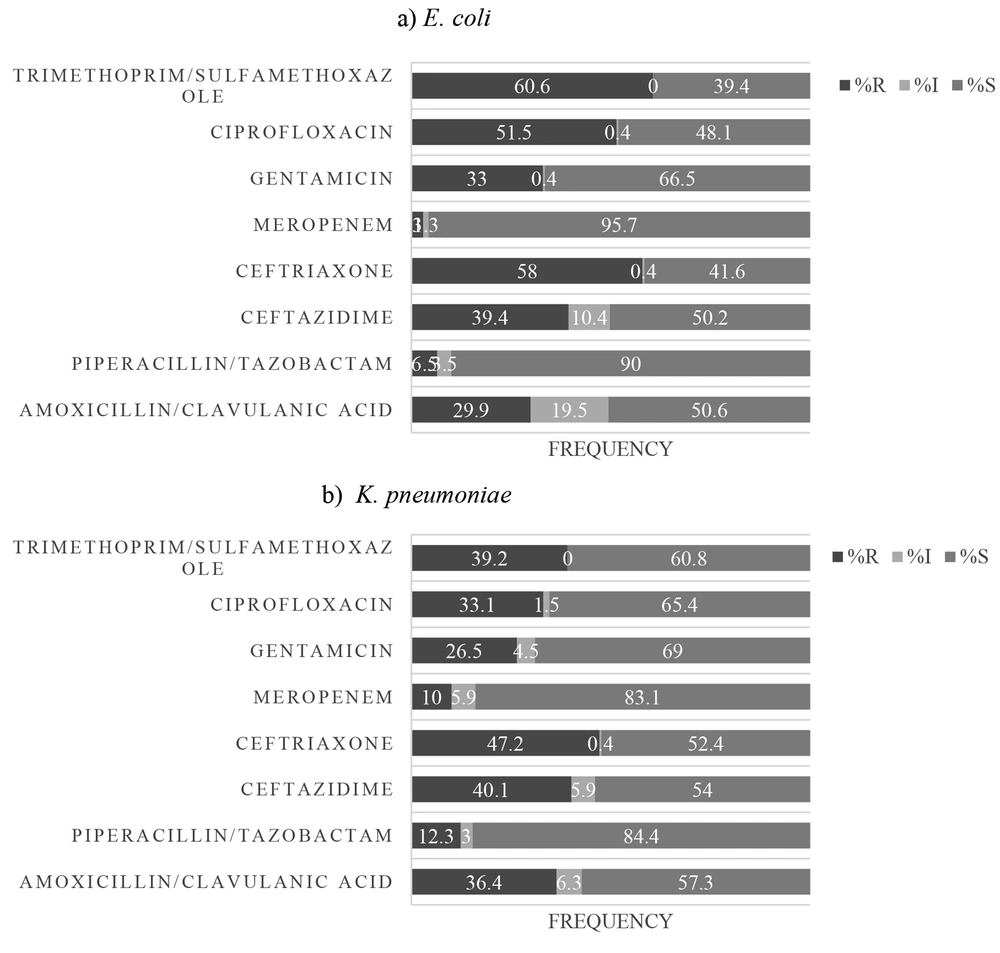
As shown in Figure 3, 60.6%, 58%, and 51.5% of E. coli isolates were resistant to SXT, CRO, and CIP, respectively. Also, 3% and 6.5% of E. coli isolates were resistant to MEM and TZP, respectively (Figure 3A). Also, 47.2%, 40.1%, 39.2%, and 36.4% of K. pneumoniae isolates were non-sensitive to CRO, CAZ, SXT, and AMC, respectively. Moreover, 10% and 12.3% of K. pneumoniae isolates were resistant to MEM and TZP, respectively (Figure 3B).
All carbapenem-resistant E. coli and K. pneumoniae isolates were ESBL-producing and resistant to CRO, CIP, MEM, CAZ, and AMP. Moreover, 33.3% of carbapenem-resistant E. coli isolates were susceptible to SXT, and 25%, 8.3%, and 4.2% of carbapenem-resistant K. pneumoniae isolates were susceptible to SXT, GEN, and TZP.
4.2. Frequency of OXA Group Genes in Escherichia coli and Klebsiella pneumoniae Isolates
In this research, 100 (43.4%) E. coli and 81 (30%) K. pneumoniae isolates contained a gene producing β-lactamases of the OXA-23-like group. In addition, nine (3.9%) and seven (2.6%) of the above isolates carried a gene encoding OXA-51-like enzymes, respectively. However, OXA-24, OXA-40, and OXA-58-like were not found in sensitive E. coli, and K. pneumoniae isolates.
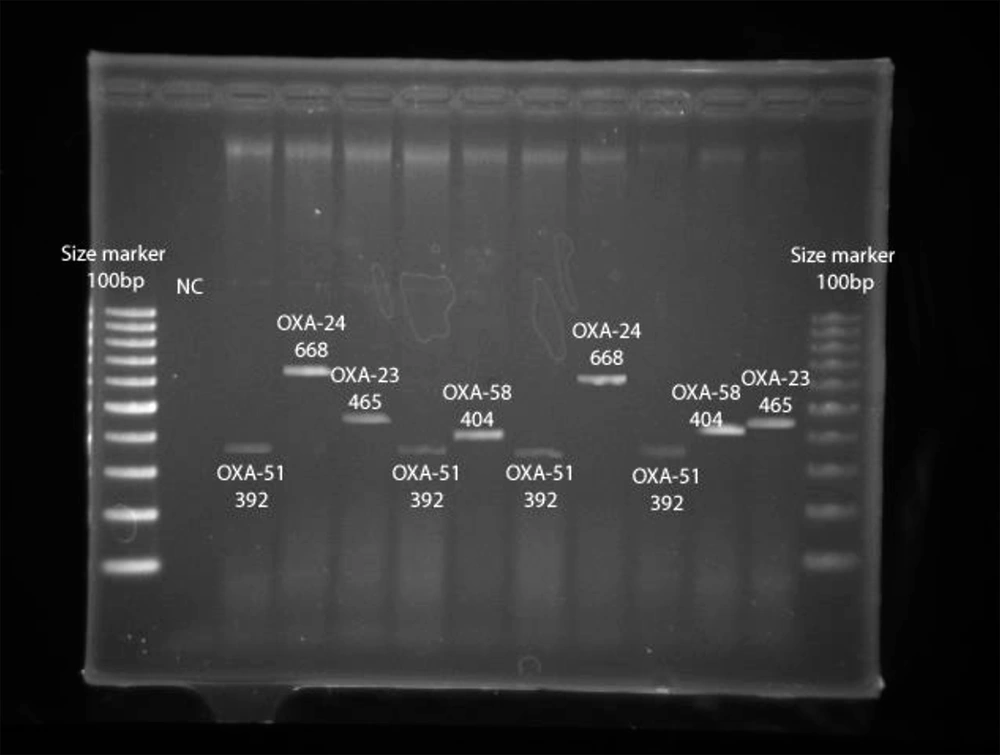
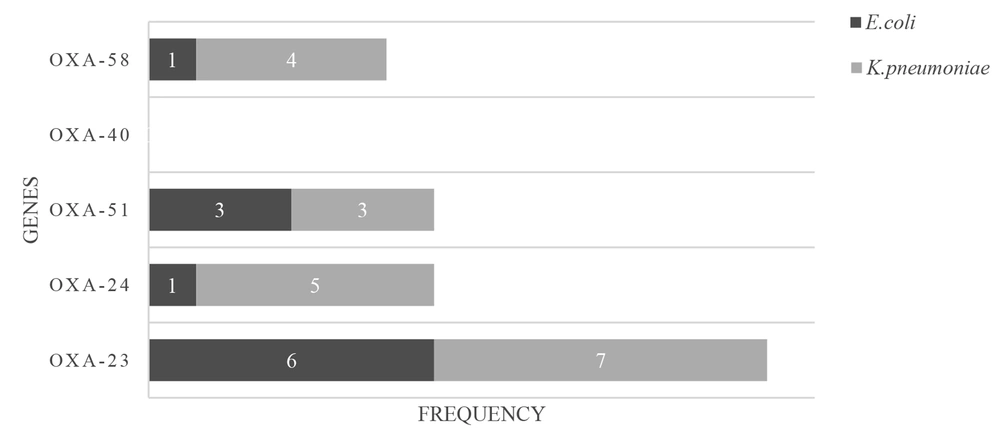
Figure 4 summarizes the presence of the OXA group genes encoding carbapenemases in carbapenem-resistant E. coli and K. pneumoniae, including OXA-23, OXA-24, OXA-40, OXA-51, and OXA-58-like groups, indicating that six (46.1%) and seven (26%) carbapenem-insensitive E. coli and K. pneumoniae isolates possessed a gene producing β-lactamase of the OXA-23-like group. Moreover, one (7.7%) and three (23.1%) of the carbapenem-insensitive E. coli isolates had a gene encoding OXA-24-like and OXA-51-like enzymes, respectively. Five (18.5%) and three (11.1%) of the carbapenem-insensitive K. pneumoniae isolates had a gene coding OXA-24-like and OXA-51-like carbapenemases, respectively.
The OXA-40-like enzyme was not found in the mentioned isolates. One (7.7%) and four (14.8%) carbapenem-insensitive E. coli and K. pneumoniae isolates had a gene encoding β-lactamase belonging to OXA-58-like enzymes (Figure 5). Also, OXA-23, OXA-24, and OXA-51-like and OXA-23 and OXA-58-like were simultaneously found in two (15%) E. coli isolates. Also, the co-existence of OXA-23 and OXA-51-like, OXA-23 and OXA-58-like, and OXA-23 and OXA-24-like was observed in three (11.1%) K. pneumoniae isolates. Different carbapenems MIC ranges were observed in E. coli, and K. pneumoniae isolates harboring the OXA-58-like gene, and these isolates showed higher carbapenem MIC ranges.
4.3. Molecular Typing of Carbapenem-Resistant Klebsiella pneumoniae and Escherichia coli
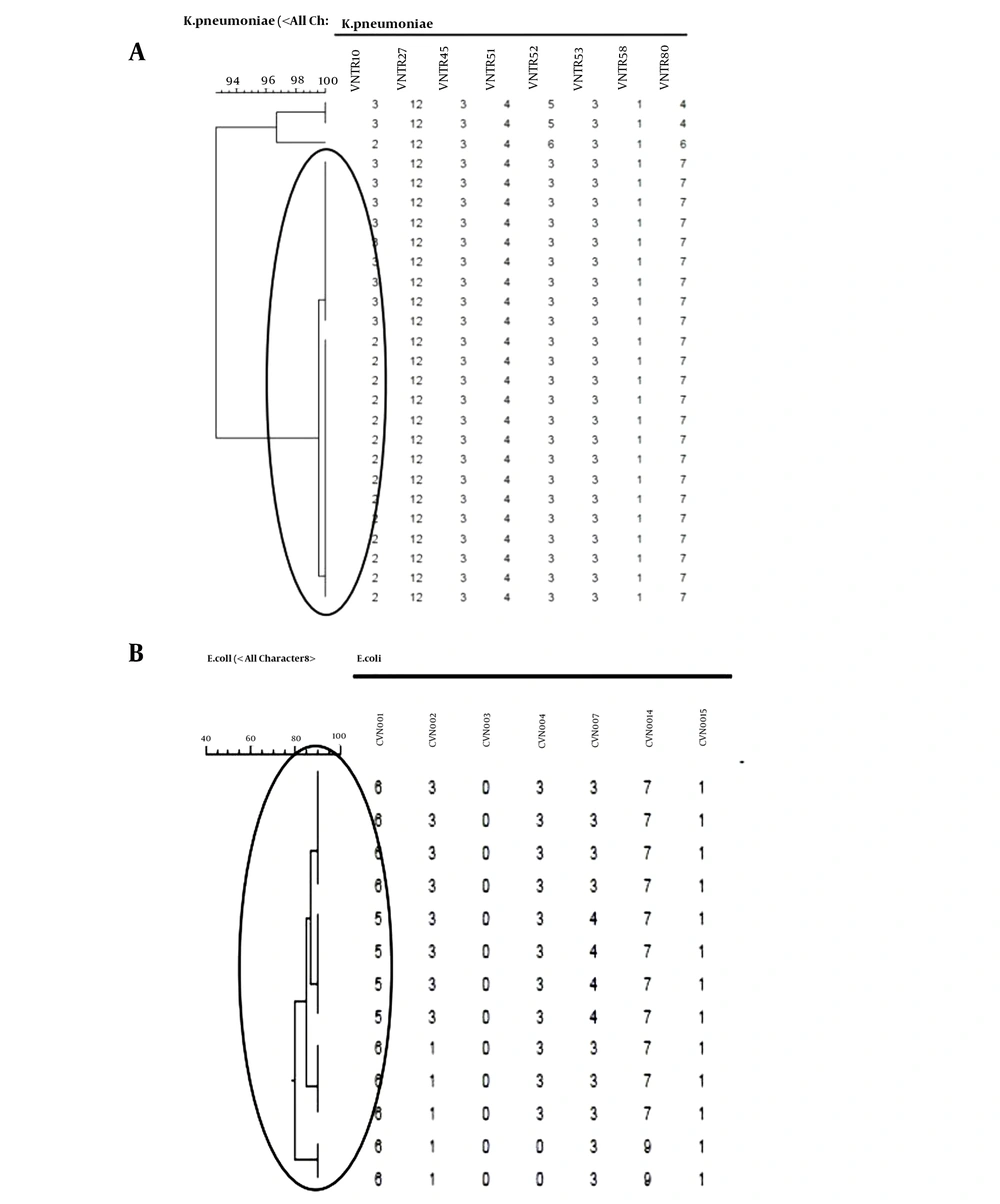
A dendrogram was formed based on MLVA for both strains using BioNumerics software ver.6.6. Figures 6A and B show that K. pneumoniae isolates were divided into three MLVA patterns and a singleton (Figure 6A), while E. coli isolates displayed two MLVA patterns (Figure 6B). According to the dendrograms, the resistance profile exhibited no significant relationship with MLVA patterns in both strains of K. pneumoniae and E. coli.
5. Discussion
In this study, in both genders and all age groups, the most common bacteria were E. coli and K. pneumoniae. The most prevalent UTIs were observed in 40 - 49 years of age group. This is probably because most patients in this sample were referred to the hospital for remedy. In a study of E. coli isolates from urine specimens in Egypt, resistance to antibiotics such as ampicillin, amoxicillin, cephalexin, and chloramphenicol was 100%. Moreover, resistance to trimethoprim-sulfamethoxazole and imipenem was 45.7% and 10.64%, respectively (23). The rates of antimicrobial resistance were slightly different. In the present study, 3% and 60.6% of E. coli isolates were resistant to meropenem and trimethoprim-sulfamethoxazole, respectively. Therefore, the location and time of study could have affected the pattern of antibiotic resistance and antibiotic use (16, 24-30). In a study of positive urine cultures for K. pneumonia in Pakistan, high resistance rates to ampicillin (100%) and trimethoprim-sulfamethoxazole (93%) were found. Also, above 85% of isolates were sensitive to carbapenem antibiotics (29). In this research, the sensitivity rate to SXT (60.8%) and MEM (83.1%) was above 60%. In addition, in this research, the sensitivity to MEM and SXT was shown to be declining, and their management must be performed with caution.
Gomes et al. detected that the multidrug resistance rate in E. coli and K. pneumoniae isolated from UTI cases was about to 86% (31). Besides, E. coli isolates displayed resistance rates of 85%, 72%, 84%, and 50% to ampicillin, trimethoprim-sulfamethoxazole, ciprofloxacin, and gentamicin, respectively, while K. pneumoniae isolates showed the insensitivity rates of 100% to ampicillin, 54% to SXT, 54% to CIP, and 27% to GEN. Also, E. coli and K. pneumoniae isolates were 100% susceptible to imipenem. In our study, resistance rates of E. coli strains were 60.6% to SXT, 51.5% to CIP, and 33% to GEN, and K. pneumoniae isolates showed the insensitivity rates of 33.1%, 39.2%, and 26.5% to ciprofloxacin, trimethoprim-sulfamethoxazole, and gentamicin, respectively. The susceptibility of E. coli and K. pneumoniae strains was 95.7% and 83.1% to meropenem as a representative of carbapenems, respectively. In Gomes et al.’s study, the resistance rates of both strains to trimethoprim-sulfamethoxazole and ciprofloxacin were more than those of our study (31). A survey on 7,098 E. coli positive cultures was done by Oteo et al. (32) in Spain that reported high resistance rates to trimethoprim-sulfamethoxazole (32.6%) and ciprofloxacin (19.3%). In our study, E. coli isolates showed higher resistance rates to trimethoprim-sulfamethoxazole and ciprofloxacin.
Comparing our results with the mentioned research indicates that diverse antibacterial resistance rates in the third world can depend on the irrational use of antimicrobial drugs, sampling biases, geographic variations, social factors, and patient characteristics. The carbapenem resistance among E. coli and K. pneumoniae isolates is a common challenge in Iran. Increasing resistance to the mentioned antibacterial drugs has made the remedy of different UTIs created by E. coli and K. pneumoniae problematic. Our study evaluated the relationship of the non-susceptibility rates of E. coli, and K. pneumoniae isolates to OXA-23-like, OXA-24-like, OXA-40-like, OXA-51, and OXA-58-like genes in Tehran. Based on the findings, all carbapenem-resistant strains were ESBL-producing and non-susceptible to CRO, CIP, MEM, CAZ, and CTX. These antibiotics can be helpful in the treatment of UTIs caused by E. coli and K. pneumoniae isolates in hospital settings.
The carbapenem resistance is mainly due to creating two β-lactamases: MBLs enzymes and oxacillinases (33). Zowawi et al. showed that the main carbapenemases in Acinetobacter baumannii (30) were carbapenem-hydrolyzing class D β-lactamases. Therefore, carbapenem-hydrolyzing class D β-lactamases were studied for E. coli, and K. pneumoniae isolates in this study. This study showed oxacillinases such as OXA-23, OXA-24 in E. coli and OXA-23 and OXA-51 in K. pneumoniae are more common in Iran. Some research has evaluated OXA group genes in E. coli and K. pneumoniae. The results of our study are consistent with the above research. Although the capability of OXA-51 and OXA-23 to hydrolyze carbapenem antibiotics is weak, the addition of the ISAba1 element upstream of the blaOXA-51/23-like gene can weaken the sensitivity rate to carbapenem antibiotics in Enterobacteriaceae (34). Budak et al. found the OXA-51-like gene in K. pneumoniae isolates in hospital samples (35). El-Hendawy et al. showed that 73.68% (14/19), 10.53% (2/19), and 21.05% (4/19) of carbapenem-insensitive K. pneumoniae isolates from hospital specimens in Egypt had OXA-48, OXA-51, and OXA-181, respectively (23).
We found significant differences in OXA group genes between carbapenem-resistant and carbapenem-susceptible E. coli and K. pneumoniae isolates. However, these differences were insignificant for the OXA-23 gene (P > 0.05). A significant association was observed between OXA group genes in E. coli and K. pneumoniae and carbapenem resistance (P < 0.05). Also, E. coli and K. pneumoniae isolates harboring the OXA-58-like gene showed higher carbapenem MIC ranges. Higher carbapenem MIC ranges indicate the role of OXA-58-like in reducing carbapenem susceptibility in the studied isolates. The OXA enzymes may become extensive quickly among Gram-negative bacteria. These gene epidemics result in the distribution of plasmids, transposons, and integrons among bacterial species. Due to the ability of integrons for the recruitment, spread, and expression of resistance genes, integrons are disseminated among Gram-negative bacteria (34, 36). Chromosomal intermediation of blaOXA-23 has been formerly demonstrated for Proteus mirabilis (37).
In Budak et al.’s research, a patient infected with multidrug-resistant A. baumannii was treated with an extended-spectrum beta-lactam and beta-lactamase inhibitor combination (35). However, two weeks later, an ertapenem-resistant K. pneumoniae isolate was discovered in the same patient. Their study suggests that the main reason for carbapenem-hydrolyzing oxacillinases is OXA-genes. Genetic events such as recombination, co-integration, and transposition in vivo (35) probably insert these genes into the chromosome. According to dendrograms (Figure 6A and 6B), carbapenem-resistant isolates possibly originated from one main clone and spread in the hospital via the vertical transmission of resistant genes. The dissemination of some genes was probably due to the transfer of mobile genetic elements.
5.1. Conclusions
Our study showed that all carbapenem-resistant E. coli and K. pneumoniae isolates were ESBL-producing and resistant to CRO, CIP, MEM, CAZ, and AMP. Also, OXA-51, 58, and 24 carbapenemases were firstly reported in the clinical strains of E. coli and K. pneumoniae isolated from volunteers with UTIs in Iran. Carbapenamases have global distribution, but there is considerable diversity at the continental, national and regional levels. A vital issue in preventing resistant strains and selecting appropriate treatment options is the awareness of the prevalence and occurrence of specific carbapenem resistance mechanisms in E. coli and K. pneumoniae. Also, OXA-23-like is the predominant gene responsible for carbapenem resistance. Further studies on large scales and in several areas are suggested for epidemiologic analyses.