1. Background
Pseudomonas aeruginosa is one of the important causes of nosocomial infections (1) and a reason for life-threatening infections among hospitalized patients (2), causing healthcare-associated infections in intensive care units (ICUs), bloodstream infections (BSI), and surgical site infections (SSI) (3). Pseudomonas aeruginosa acquires resistance genes encoding extended spectrum beta-lactamases (ESBLs) enzymes, which hydrolyze penicillins and cephalosporins, including third and fourth-generation cephalosporins and aztreonam (ATM) (4). Extended spectrum beta-lactamases are classified into two structural Ambler classes, A (such as VEB, BEL, TEM, SHV, CTX-M-type, and PME) and D (such as the OXA family) (4, 5). These genes are rapidly spreading in the world, posing the serious threat of treatment-resistant P. aeruginosa infections and subsequently, increased rates of morbidity and mortality (6, 7). The epidemiological subtypes of P. aeruginosa strains are determined using different genotyping methods (7). Pulse field gel electrophoresis (PFGE), repetitive element sequence (Rep)-PCR, and enterobacterial repetitive intergenic consensus (ERIC)–PCR are the most widely used methods for genotyping of these isolates (8).
Several studies have been published on the nosocomial spreading of ESBL-producing P. aeruginosa isolates (9-11). There is a worldwide concern over the propagation of resistant clones of this microorganism in hospitals. Over decades, multiple reports have warned of epidemic outbreaks caused by multiple-drug resistant (MDR)/extensively-drug resistant (XDR) strains within the hospital environment. More recently, evidence has been provided on the existence of MDR/XDR global clones disseminating in several hospitals worldwide, known as high-risk clones [6, 7]. Therefore, identifying the source of these bacteria, detecting their strains, and characterizing their various resistance mechanisms are of great importance and significance. Considering the above-mentioned, studying different aspects of P. aeruginosa seems to be necessary.
2. Objectives
In this study, we aimed to determine the genotypes of ESBL-producing P. aeruginosa isolated from patients’ specimens in Kurdistan’s teaching hospitals using phenotypic and molecular methods.
3. Methods
3.1. Bacterial Isolates
One hundred forty six clinical isolates of Pseudomonas spp. were obtained from microbiology laboratories of five tertiary referral hospitals. These strains had been isolated from the specimens obtained from different patients (each isolate was from a unique patient). All patients with Pseudomonas infections (nosocomial or non-nosocomial) were included in this research. Nosocomial infections occur after 48 - 72 hours of hospitalization (12).
3.2. Pseudomonas aeruginosa DNA Extraction
The DNA samples used in Rep-PCR and ERIC-PCR were extracted using a Gram-negative DNA extraction kit (SinaClon, Iran).
3.3. Pseudomonas aeruginosa Detection by PCR
The gyrB (Gyrase B), as a housekeeping gene, was used for identifying P. aeruginosa. PCR was performed using (F)-5`-CCTGACCATCCGTCGCCACAAC-3` and (R)-5`-CGCAGCAGGATGCCGACGCC-3`) primers (222 bp) (SinaClon, Iran). The positive control was P. aeruginosa strains ATCC 25922 (Pasteur institute, Iran) (13).
3.4. Phenotypic Identification of ESBL-Producing Pseudomonas aeruginosa Strains
Mueller-Hinton agar (MHA) (HiMedia, India) was used to conduct the disk approximation test. A disk containing amoxicillin/clavulanic acid (AMC) (20/10 µg) was placed in the center of the plate, and cefpodoxime 10 μg (CPD), cefepime 30μg (FEP), CAZ, and CTX were placed 2.5 cm away from AMC. For CDT, ceftazidime-clavulanate (CAZ/C) (30/10 µg), CAZ, CTX/C (30/10 µg), CTX, CPD/C (10μg/1μg), and CPD were placed 2.5 cm apart from each other all over MHA. The plates were incubated at 37°C for 24 hours, and the clear zone of inhibition was measured. The results were compared to the standards published by the clinical and laboratory standardization Institute (CLSI) (14).
3.5. Molecular Detection of Class A Beta-lactamase Genes and Genotyping
Specific primers (SinaClon, Iran) were used in this study to detect the blaSHV and blaTEM genes by PCR (15). Positive controls were P. aeruginosa strains expressing blaTEM and blaSHV (Pasteur Institute, Iran), and deionized water was used as the negative control. Also, Rep-PCR and ERIC-PCR were performed for 56 strains isolated from patients with nosocomial infections. Rep-PCR applying ERIC-PCR primers (Macrogen, South Korea) was used for genotyping P. aeruginosa (16).
3.6. Pulsed-field Gel Electrophoresis (PFGE) for Genotyping
In this step, 56 P. aeruginosa isolates from patients with nosocomial infections were selected for PFGE analysis and genotyping using the XbaI restriction enzyme (Fermentas, USA). Electrophoresis in a PFGE system (Chef Mapper; Bio-Rad Laboratories, Hercules, CA, USA) was performed at Kermanshah University of Medical Sciences, Iran (17).
3.7. Statistical Analysis
Data were analyzed using Stata software and statistics such as frequency and Fisher’s exact test. The logistic regression model was used to test the impact of independent variables in the presence of each of other variables (P ≤ 0.05). The analysis of the banding patterns of Rep-PCR, ERIC-PCR, and PFGE was performed by Gel.J V.1.3 software using the dice tolerance of 2.0, the UPGMA method, and 95% similarity for drawing dendrograms.
4. Results
4.1. Phenotypic and Molecular Identification of Pseudomonas aeruginosa
The isolates were confirmed as P. aeruginosa by PCR and phenotypic testing, delivering the accuracies of 91.78% and 91.09%, respectively.
4.2. Clinical Specimens’ Data
Pseudomonas aeruginosa strains were isolated from the clinical samples gathered from Toohid (95; 70.89%), Besat (33; 24.62%), Imam Hossein (4; 2.98%), Imam Khomeini (1; 0.7%), and Fajr (1; 0.7%) hospitals in Kurdistan province.
4.3. Disk Approximation Test and CDT Results for ESBL
The disk approximation test proved that 24 of 134 (17.91%) P. aeruginosa isolates were ESBL producers. Sanandaj (23; 95.83%) and Bijar (1; 4.16%) were the cities where most ESBL-producing P. aeruginosa strains were isolated from. Regarding CDT, 84/134 (62.68%) P. aeruginosa isolates were ESBL producers.
4.4. PCR Results for bla<sub>SHV</sub> and bla<sub>TEM</sub> Genes
Out of 24 P. aeruginosa isolates that were ESBL producers according to the disk approximation test, one (4.16%) and 14 (58.33%) were found to express the blaSHV and blaTEM genes, respectively. Based on the disk approximation test, out of a total of 110 non-ESBL-producing isolates, seven (6.36%) and 75 (68.18%) carried the blaSHV and blaTEM genes, respectively (Tables 1 and 2).
| Disk Approximation Test | CDT | |||||
|---|---|---|---|---|---|---|
| Yes | No | Total | Yes | No | Total | |
| PCR for blaSHV gene | ||||||
| No | 23 (18.52) | 103 (81.74) | 126 | 78 (61.90) | 48 (38.09) | 126 |
| Yes | 1 (12.50) | 7 (87.50) | 8 | 6 (75) | 2 (25) | 8 |
| Total | 24 (17.91) | 110 (82.08) | 134 | 84 (62.68) | 50 (37.31) | 134 |
| PCR for blaTEM gene | ||||||
| No | 10 (22.22) | 35 (77.77) | 45 | 26 (57.77) | 19 (42.22) | 45 |
| Yes | 14 (15.73) | 75 (84.26) | 89 | 58 (65.16) | 31 (34.83) | 89 |
| Total | 24 (17.91) | 110 (82.08) | 134 | 84 (62.68) | 50 (37.31) | 134 |
Frequency of ESBL-Producing Pseudomonas aeruginosa Carrying blaSHV and blaTEM PCR for blaSHV Gene a
| Type of Infection | P. aeruginosa Strains | P. aeruginosa Producing ESBL (Disk Approximation Test) | P. aeruginosa Non-producing (CDT) |
|---|---|---|---|
| Non-nosocomial | 78 (58.20) | 16 (66.66) | 50 (59.52) |
| Nosocomial | 56 (41.79) | 8 (33.33) | 34 (40.47) |
| Total | 134 | 24 | 84 |
Frequency of ESBL-Producing Pseudomonas aeruginosa Isolates in Samples Obtained from Patients with Nosocomial and Non-nosocomial Infections a
4.5. The Results of ERIC-PCR Genotyping
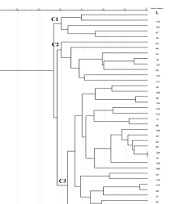
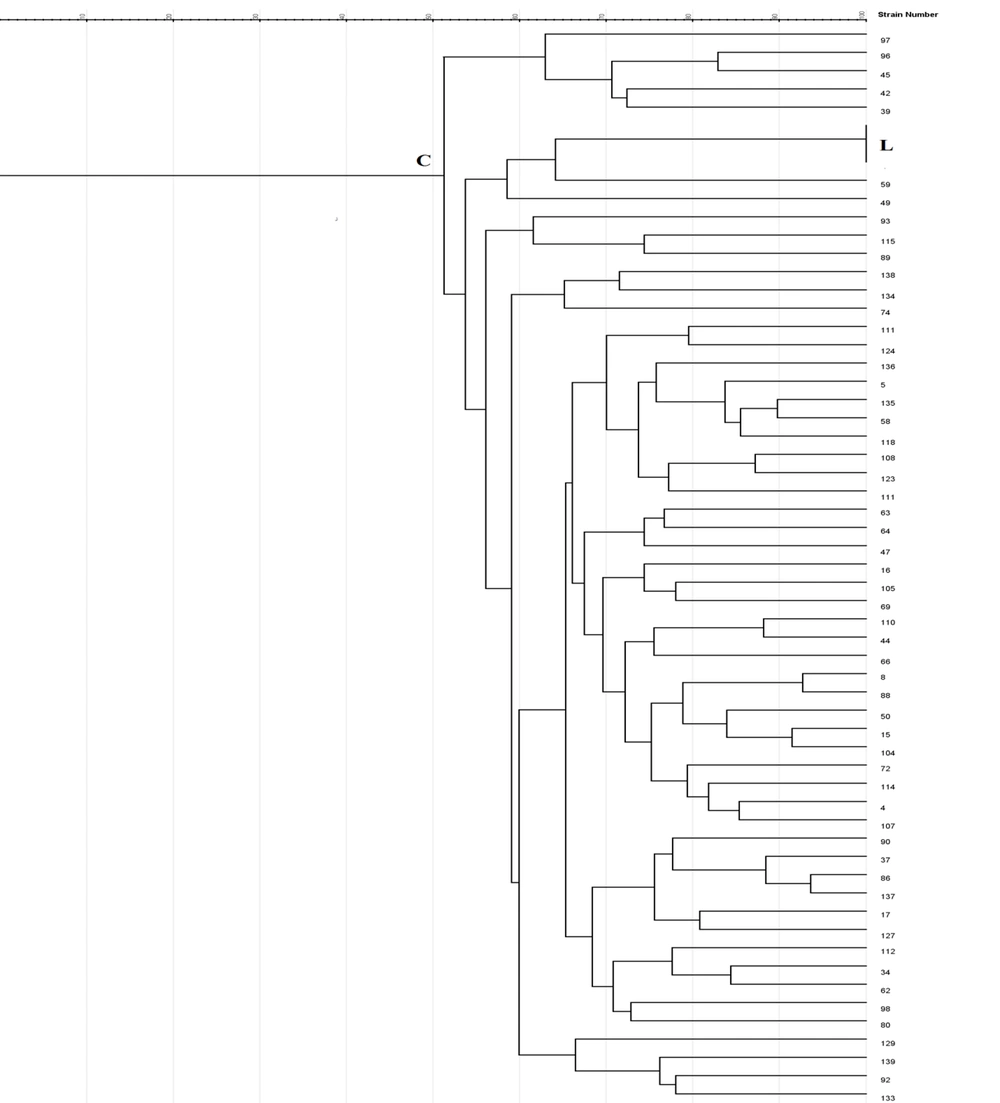
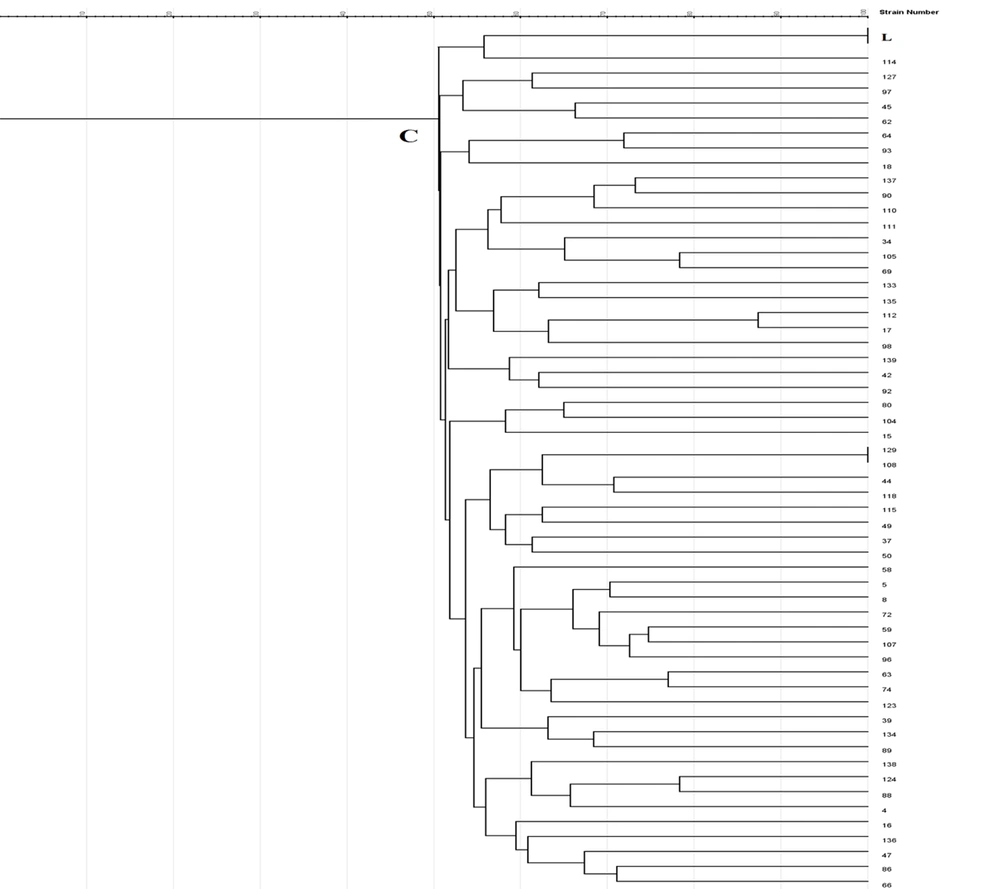
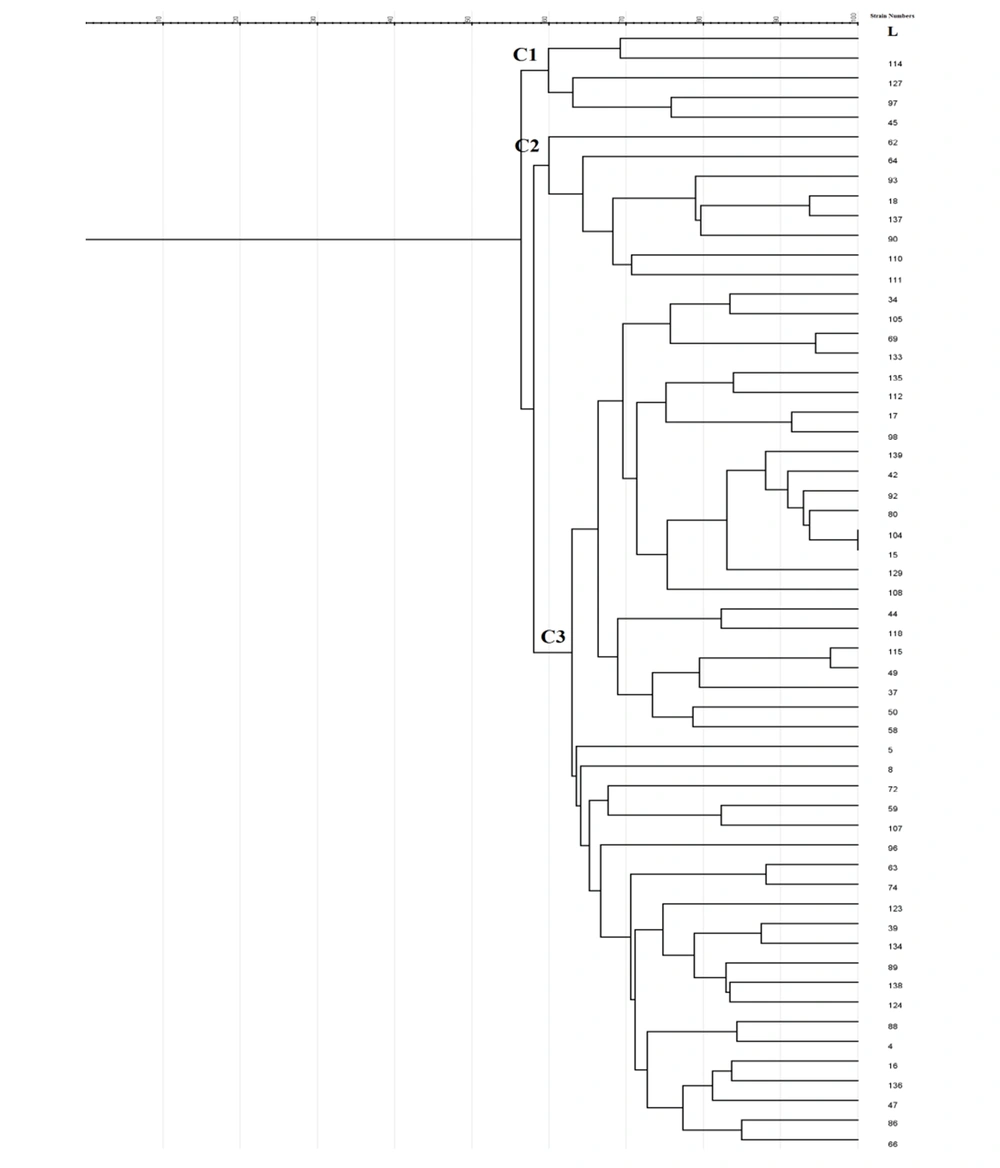
Enterobacterial repetitive intergenic consensus -PCR showed 56 different patterns with 5 - 14 bands ranging from 100 to 3000 bp in all 56 isolates. One main cluster (C) with 51% similarity was identified. No similar clonality pattern was observed among the isolates obtained from nosocomial infections. All 56 isolates were included in cluster C, which was the predominant ERIC profile (Figures 1 - 3).
4.6. The Results of Rep-PCR Genotyping
In Rep-PCR, bands with approximate sizes between 100 and 1500 bp were observed. Based on the results of Rep-PCR clustering, isolates from nosocomial infections with 51% similarity were classified into 55 groups. All strains belonged to the cluster C (Figures 1 - 3).
4.7. The Results of PFGE Genotyping
According to PFGE analysis, 56 isolates with 60% similarity obtained from nosocomial infection samples were classified into 55 pulsotypes. In the cluster C3, two strains (104 and 15) showed 100% genetic similarity. The C3 cluster was the dominant pulsotype, harboring most of the isolates (Figures 1 - 3).
4.8. Statistical Findings
Statistical analyses showed no association between the presence of blaSHV and blaTEM genes and ESBL production by P. aeruginosa isolates (P ≥ 0.05). Also, blaSHV and blaTEM expression by P. aeruginosa strains showed no significant association with age, sex, city of living, inpatient/outpatient status, hospital wards, and specimen’s type (P ≥ 0.05). However, blaSHV and blaTEM positivity was significantly associated with ICU admission and tracheal samples (P ≤ 0.05). No significant association was observed between ESBL production or expression of blaSHV and blaTEM and ERIC, Rep-PCR, and PFGE patterns (P ≥ 0.05).
5. Discussion
Pseudomonas aeruginosa exists in different environments and causes wide-spectrum infections in individuals. Therefore, surveillance of P. aeruginosa strains and their transmission among patients in hospitals seems highly necessary (12). The rate of nosocomial infections differs in various geographical regions (18-20), which may be due to health system policies and study conditions. In our study, a significant relationship was observed between ICU admission, tracheal samples, and nosocomial infections. The patients admitted to the ICU stay more time in hospitals, and therefore, are more prone to acquired infections.
Pseudomonas aeruginosa is an opportunistic bacterial pathogen, and a common cause of infection in people admitted to the ICU. A study reported that the prevalence of blaSHV-12 and blaTEM-24 among ESBLs were as 17.6% and 20.5%, respectively (21). These rates were higher than the blaSHV-12 and blaTEM-24 positivity rates observed in our study. In the present research, the disk approximation test proved that out of 17.91% of ESBL-producing P. aeruginosa isolates, 4.16% and 58.33% were positive for blaSHV and blaTEM, respectively. Among non-ESBL-producing isolates, 6.36% and 68.18% expressed the blaSHV and blaTEM genes, respectively. According to CDT, 62.68% of P. aeruginosa isolates were ESBL producers, of whom 7.14% and 69.04% expressed the blaSHV and blaTEM genes, respectively. On the other hand, 4% and 68% of the isolates that were negative for ESBLs based on CDT harbored the blaSHV and blaTEM genes, respectively.
Compared to molecular methods, disk phenotypic methods provide a simple and inexpensive strategy that can be easily used in clinical laboratories. Beta-lactamase genes can be transferred among different species of bacteria via various gene transferring mechanisms, for example, via plasmids and transferable elements. Also, genotype will not always reflect the phenotype, and mutations and environmental factors will affect this relationship (22). In a study in Iran in 2017, out of 75 P. aeruginosa isolates, 20 common types and 20 unique patterns were identified using Rep-PCR (16). The clonality of metallo-beta-lactamase (MBL)-producing isolates was assessed by ERIC-PCR, revealing three genotypes, including type A (76.8%), type B (20.3%), and type C (2.9%) (12). In the present study, genotyping by ERIC-PCR, Rep-PCR, and PFGE showed 56, 55, and 55 different patterns. Only two isolates assessed by PFGE and Rep-PCR showed a 100% genetic similarity.
In the present study, Rep-PCR showed that two samples (No. 129 and 108, one from Marivan, Fajr hospital, Men’s internal ward, isolated from urine and the other from Snandaj, Toohid hospital, the emergency ward, isolated from blood) had 100% genetic similarity. Also, a 100% similarity was observed between samples No. 104 and 15 by PFGE (both from Sanandaj, Toohid and Besat hospitals, both isolated from patients admitted to the ICU and both from tracheal samples. All four samples were ESBL producers according to CDT, but no genes were detected in any of these. It is likely that these bacteria have been transferred between hospital staff and patients or from patients in one hospital to another hospital (i.e., circulating strains). Various epidemiological studies have reported completely similar strains, suggesting that these samples might have had a common source. Our results showed diversity in terms of resistance among P. aeruginosa strains in the Kurdistan providence of Iran.
Among molecular epidemiology techniques, PFGE is a gold standard for characterizing pathogenic bacteria (23). In this study, only 56 out of 134 isolates were analyzed by PFGE. Using PFGE, 55 pulsotypes were detected. The pulsotypes of C3 were dominant, and most of the isolates belonged to this cluster, in which two samples (No. 104 and 15) had a 100% genetic similarity. Overall, PFGE provides a more discriminatory and powerful method to identify the isolates involved in nosocomial infections. The main disadvantage of this method is its low consistency between isolates analyzed in different studies. Review studies have reported similar results (9, 11, 24). Therefore, PFGE is one of the most robust methods for molecular epidemiology analysis and fingerprinting of bacterial isolates.
5.1. Conclusions
In this study, heterogeneity was observed among clusters, and no particular source could be cited for these isolates. In the current study, a significant ratio of P. aeruginosa isolated from hospitalized patients and outpatients were ESBL producers, indicating the widespread use of beta-lactam antibiotics and the high rate of resistance among these isolates. Therefore, efficient and sustained controlling measures and proper antibiotic policies should be implemented in hospitals.