1. Background
The number of lives at risk due to the exponential increase in antimicrobial resistance is predicted to reach 10 million by 2050, with 100 trillion United States dollar annual loss of the global economy (1). To date, resistance against approximately all known antibiotics has emerged and is considered a severe global threat (2). Antibiotic resistance is currently acknowledged as a global medical concern that has endured more than 70 years as a result of continuous extensive antibiotic abuse without appropriate surveillance and diagnostics (3, 4). Consequently, multidrug-resistant (MDR) infections are considered a leading health hazard and declared as a significant public health problem by world healthcare agencies, such as the United States Centers for Disease Control and Prevention, the European Center for Disease Prevention and Control, and the World Health Organization (WHO) (5).
Klebsiella Pneumoniae is a common pathogen in hospital settings responsible for several hospital-acquired infections (HAIs) (6). It is a Gram-negative nonmotile bacteria with a relatively large genome and metabolic flexibility that allows it to vastly associate with human diseases (7). Classical K. pneumoniae (cKp) and hypervirulent K. pneumoniae (hvKp) are two prominent phenotypes of K. pneumoniae predominantly associated with various hospital- and community-acquired infections (CAIs) (8). The cKp is a leading opportunistic pathogen associated with HAIs, and its common clinical manifestations are pneumonia, urinary tract, and wound infections, any of which can progress to bacteremia (9). Patients infected with hvKp and liver abscess can also develop metastasis, which leads to severe infections of the eye, lung, and central nervous system. Other common clinical manifestations associated with hvKp infections included epidural abscess, severe skin, soft tissue, and bone infections (eg, necrotizing fasciitis). Furthermore, hvKp is commonly associated with CAIs (10, 11). Consequently, in 2017, the WHO prioritized K. pneumoniae as a globally disseminated MDR pathogen with epidemic potential (12).
Clinically, it has been observed that K. pneumoniae is becoming resistant to every class of antibiotics. The fundamental factor for resistance in K. pneumoniae is its ability to horizontally acquire resistant genes (3). However, intrinsically K. pneumoniae is resistant to aminopenicillins by producing class A β-lactamase encoded by chromosomal gene blaSHV (13), the variants of which when acquired by K. pneumoniae produce extended-spectrum β-lactamase activity (conferring resistance to third-generation cephalosporins) and occasionally even carbapenemase activity (14). Resistance to antibiotics, such as carbapenems, fluoroquinolones, chloramphenicol, tigecycline, and aminoglycosides, is associated with efflux pump activity (15). Consequently, antibiotic therapeutic choices against resistant K. pneumoniae remain limited; nevertheless, other measures, such as phage therapy, have been considered alternative measures to prevent and control infections caused by MDR K. pneumoniae (16).
The term bacteriophage was first introduced by Felix d'Herelle in proving the statement by William Twort that the clear zones in bacterial cultures are due to viruses (17). It has been demonstrated that the lysis ability of bacteriophages is more suitable for the efficient reduction of bacterial growth. Along with the lytic ability, lytic phages have a short latent period, compared to lysogenic phages, and do not produce any toxins or are not responsible for the dissemination of resistant genes (18). The bacterial cell lysis is carried out by different phage proteins, such as holins, endolysins, and spanins, as they play a role in assembly bursting out of phage from bacteria. The degradation of peptidoglycan is carried out by endolysins; however, cell membrane disruption occurs through spanins and holins (19). The prevalence of MDR K. pneumoniae is reported from Pakistan, and the isolated strains harbored the globally disseminated resistance genes, including blaOXA-48, blaNDM, and blaKPC-2 (20, 21). Recently novel MDR K. pneumoniae sequence type ST147 has been discovered; the strain was resistant to most antibiotics and harbored blaOXA-48, blaNDM, and blaKPC-2 (22). This increase in antibiotic resistance and emergence of MDR, extremely drug-resistant (XDR), and pandrug resistant pathogens, especially K. pneumoniae, demands an alternative approach to overcome antibiotic resistance as suggested by the WHO.
2. Objectives
Phage therapy has been proven to be the most efficient method to overcome antibiotic resistance. Previously, we have reported the efficacy of phage-antibiotic combination therapy against MDR K. pneumoniae (23). However, this study reports the occurrence of hvKp and the lytic potential of bacteriophage against the isolates that alone demonstrated very promising results and significant lytic and stability patterns.
3. Methods
3.1. Isolation of K. pneumoniae
The samples were collected from different hospitals in district Faisalabad, Pakistan. The specimen sources included pus exudates, tracheal aspirate, sputum, wound swabs, and catheters tips. The samples containing cotton swabs and containers were tightly closed and wrapped in sterile strip plastic bags to avoid contamination (24). All the samples were cultured on MacConkey agar, and overnight incubation was given at 37°C. The single colony of bacterial growth from MacConkey agar was then streaked over on K. pneumoniae selective base agar (HiCromeTM Klebsiella selective agar M1573-500G) and incubated at 37°C for 24 hours. The freshly grown pure colonies of K. pneumoniae were then used to determine the phage host range (25).
3.2. Biochemical Characterization and Microscopy
The isolates were identified using standard microbiology procedures followed by biochemical characterization through API 20E kit (BioMérieux, France) (26).
3.3. Phenotypic Characterization of Hypervirulent K. pneumoniae
The phenotypic analysis of hypermucoviscosity was performed by string test. Newly grown colonies were touched at the center with a sterile wire loop, and the loop was moved upward to produce a string from the colony. The string length > 5 mm was considered a positive string test (27).
3.4. Antimicrobial Susceptibility Testing
Antibiotic susceptibility test (AST) was performed using the previously described Kirby-Bauer Disc diffusion method (28). Briefly, the overnight grown culture of K. pneumoniae at 0.5 McFarland standard was spread over Mueller-Hinton agar (MHA). Antibiotic disks were placed on MHA containing bacterial culture, and the plates were incubated at 37ºC for 24 hours. For positive control, previously determined MDR K. pneumoniae NCBI GenBank accession number of MF953600 was used for AST. Antibiotic susceptibility to ampicillin (30 μg), amoxicillin (30 μg), ceftriaxone (30 μg), cefoxitin (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), cefepime (30 μg), amikacin (30 μg), gentamicin (30 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), colistin (10 μg), imipenem (10 μg), and doripenem (10 μg) was determined. The results were interpreted based on the Clinical and Laboratory Standards Institute (2020) guidelines (29).
3.5. Isolation of Bacteriophage
Bacteriophages were isolated from the hospital wastewater. The wastewater was collected from different hospitals of district Faisalabad. Briefly, the hospital wastewater was collected in locally purchased 500 mL plastic bottles. All plastic bottles were then tightly closed and wrapped in sterile bags (strip plastic bags) to avoid contamination (30). All bottles were placed on the benchtop to allow the large debris to settle down for 1 to 3 hours. The collected wastewater was treated with 1% chloroform to kill bacteria filamentous temperate phages and facilitate the detachment of phages from bacteria (31). The wastewater was then subjected to the bacteriophage enrichment process.
3.6. Bacteriophage Enrichment and Purification Assay
Bacteriophage enrichment and purification were performed as previously described (32). Briefly, 90 mL of wastewater samples, along with 1 mL of bacterial broth, was suspended in (10X) 9 mL Luria-Bertani (LB) medium and incubated for 24 hours at 37ºC. The overnight culture was then subjected to centrifugation at 12000 rpm for 15 minutes. For plaque assay, the double agar overlay method was used. The centrifuged supernatant was then filtered through a 0.22 um pore filter. The filtered supernatant was then diluted in 900 µL of phosphate-buffered saline (PBS), along with 100 µL of K. pneumoniae, then added to 0.5% LB medium-soft agar. The soft agar containing phage host suspension was poured onto the LB agar plate and incubated at 37ºC for 24 hours. After incubation, the single plaque was picked with a sterile tip processed through the aforementioned process for phage purification (33).
3.7. Host Range Determination
The host range of isolated bacteriophages was determined by spot test as previously described (34). Briefly, overnight bacterial culture at 0.5 MacFarland standard was used to make lawns over MHA. The unfiltered centrifuged phage suspension previously obtained from the enrichment assay was utilized to make spots on freshly spread bacterial lawns. Moreover, 5 µL of phage suspension was spotted over bacterial lawns and incubated for 24 hours at 37ºC. In addition to the K. pneumoniae isolates, some other bacteria, including Escherichia coli, Pseudomonas aeruginosa, and Salmonella typhi, were subjected to spot tests to determine the host range. However, K. pneumoniae remained the test host bacterium for the study. After 24 hours of incubation, the clear zones between bacterial growth due to phage lytic activity were determined. The paired t-test was performed with 95% T alpha half for statistical analysis.
3.8. Phage Stability Test
Isolated bacteriophage stability was tested against different environmental conditions, including temperature, pH, and chloroform. The thermal stability of bacteriophage was determined against different temperatures, including 0, 4, 15, 37, 48, and 55ºC (35). Isolated bacteriophage was incubated at various ranges of temperature at 108 PFU/mL for 4 hours. After incubation, the phage titer was determined by the double agar overlay method. For the determination of pH stability, first, the pH of PBS was adjusted to a variety of 2, 4, 7, 9, and 11 by adding 1 M sodium hydroxide and hydrochloric acid. In addition, 108 PFU/mL of phage suspension was incubated for 4 hours at various pH ranges, and phage titer was determined by the double agar overlay method (36). Then, 10, 15, and 20 µL of chloroform were mixed with 990, 985, and 980 ul of PBS. Furthermore, 100 µL of phage suspension at a 106 PFU/mL concentration was suspended in 900 ul PBS with various chloroform concentrations using 1.5 mL Eppendorf tubes. The tubes were incubated at 37ºC for 4 hours, and the titer of surviving phages was determined as PFU/mL (35).
3.9. One-Step Growth Curve
The one-step growth curve was determined as previously described (32). The particular experiment was conducted to determine the phage host interaction and some important steps during phage infection, such as adsorption period, latent period, and burst size. Briefly, initially, an overnight culture of K. pneumoniae in LB was adjusted to 108 CFU/mL. For phage adsorption, 0.9 mL of bacterial suspension was mixed with 0.1 mL of the phage (multiplicity of infection [MOI] = 0.01) and incubated at 37ºC for 5 minutes. The mixture was diluted, and the samples (0.1 mL) were taken at 5-minute intervals up to 30 minutes, followed by 15-minute intervals up to 90 minutes, to determine the number of phages by the double-layer agar method (32). The latent period, burst time, and burst size were calculated from the one-step growth curve.
3.10. Phage Adsorption Assay
An overnight culture of susceptible K. pneumoniae in Mueller-Hinton broth was adjusted to 108 CFU/mL. The equal volumes of the bacterial culture and phage suspension at (106 PFU/mL) were mixed and incubated at 37ºC for 5 and 10 minutes, respectively. The cultures were centrifuged at 10000 × g for 5 minutes and filtered through a 0.22 μm filter, and the numbers of free phages were determined using the double-layer agar method (32). The reduction in phage titer showed the number of phages adsorbed to the bacterial cells (37).
4. Results
4.1. Distribution of K. pneumoniae
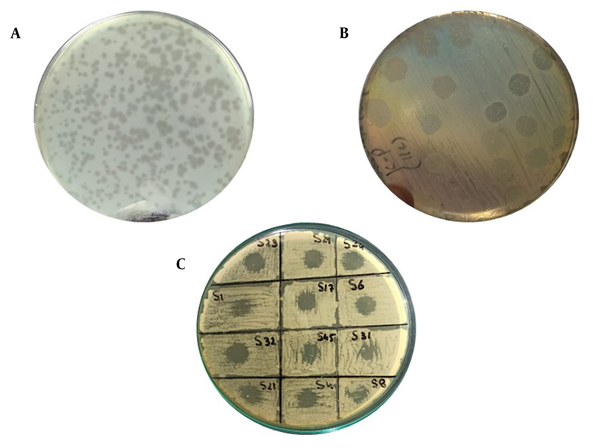
Among 50 clinical samples, 22 isolates (44%) were identified as K. pneumoniae. Out of 22 isolates, 11 isolates (50%) were detected as hypervirulent/hypermucoid K. pneumoniae as the string length was observed higher than 5 mm (Figure 1). Overall, out of 11 hvKp isolates, 3 isolates (27%) were from wound samples and 3 isolates (27%) were positive from catheters tips and pus each. Only 1 isolate (9%) was detected as hvKp from tracheal aspirates; however, no hvKp isolate was detected from sputum (Table 1).
A, Pink mucoid colonies of K. pneumoniae on MacConkey agar; B, Antibiotic susceptibility test results on Mueller-hinton agar plates by disk diffusion method (using antibiotic disks and Clinical and Laboratory Standards Institute guidelines [2020] for K. pneumoniae); C, String test showing the confirmed hypervirulent Klebsiella pneumoniae isolates; D, Comparative analysis of resistance against different antibiotics (maximum resistance against ceftriaxone in gray and minimum resistance against colistin in blue)
| Sample Sources | Total Collected Samples | Isolates | Multidrug Resistance Out of Positive Isolates | Hypervirulent K. pneumoniae Out of Positive Isolates | Phage Susceptibility of Multidrug-Resistant Isolates |
|---|---|---|---|---|---|
| Pus | 10 | 5 (50) | 4 (80) | 3 (27) | 4 (100) |
| Infectious wounds | 9 | 5 (55) | 4 (80) | 3 (27) | 3 (75) |
| Catheters tips | 12 | 5 (41) | 3 (60) | 3 (27) | 2 (66) |
| Sputum | 8 | 3 (37) | 1 (33) | 0 | 1 (100) |
| Tracheal aspirates | 11 | 4 (36) | 2 (50) | 2 (18) | 2 (50) |
| Total | 50 | 22 (44) | 14 | 11 (50) | 12 |
Distribution of Klebsiella pneumoniae (ie, Host Bacterium) Isolates with Respective Phage Susceptibility a
4.2. Antibiotic Susceptibility Testing
Overall, out of 22 confirmed isolates, 14 isolates (64%) were detected as MDR. Moreover, 4 isolates (28%) were MDR K. pneumoniae recovered from pus and wound samples each followed by catheter tips (n = 3; 21%), tracheal aspirates (n = 2; 14%), and sputum (n = 1; 7%), respectively. Additionally, out of 11 hvKp isolates, 5 isolates (45%) were confirmed as MDR K. pneumoniae. Nevertheless, AST revealed maximum and minimum resistance against ampicillin (86%) and colistin (22%) detected in the isolates of the study, respectively. Figure 1 illustrates the detailed resistance profiling of the isolates.
4.3. Phage Purification
For purification, the double agar overlay method was used. Clear plaques were observed on the bacterial lawn. The plaque size of isolated bacteriophages ranging from 0.5 - 2.0 mm was recorded. The average plaque formation time was estimated at 8±1 hours.
4.4. Host Range
The host range was determined through the spot test indicating that the isolated phage specifically lysed K. pneumoniae; nonetheless, no lytic activity was determined against other bacteria, including E. coli and P. aeruginosa. Overall, the phage showed activity against 16 (77%) out of 22 K. pneumoniae isolates (P = 0.003; Table 2). However, among 14 MDR K. pneumoniae isolates, 12 isolates (85%) were susceptible to the isolated bacteriophages, among which 5 isolates (35%) were hvKp (Table 1 and Figure 2).
| Pearson Correlation | df | Mean Difference | Standard Deviation | Standard Error | T Alpha Half 95% CI | Lower CI | Upper CI | P-Value |
|---|---|---|---|---|---|---|---|---|
| 0.784 | 4 | 2 | 0.707107 | 0.316228 | 2.7764 | 1.122025 | 2.354816 | 0.003198 |
Statistical Analysis (Paired T-Test) of Data Obtained from Phage Host Range
4.5. Bacteriophage Stability
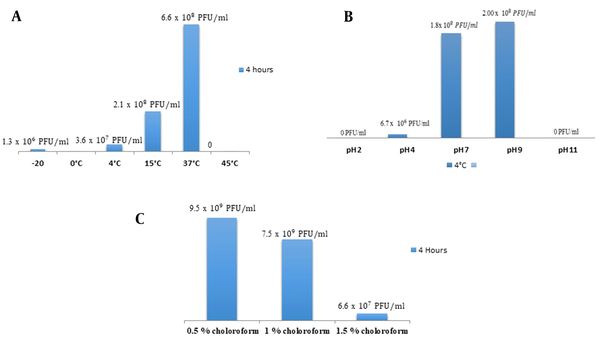
The isolated bacteriophage showed a broad host range and was tested for its stability against various environmental conditions. No plaques were observed at pH 2; however, few plaques were detected at pH 4. The high titer of plaques was calculated at pH 7. At pH 9, the significantly higher titer of plaques was estimated. However, no plaques were observed at pH 11. In case of thermal stability, no plaques were observed at 0ºC. The phage showed significant lytic activity at 37ºC as maximum phage titer was observed at this temperature. Isolated phage remained stable at 0.5 - 1.5% chloroform treatment and showed lytic activity (Figure 3).
A, Comparative analysis of phage titer at various ranges of temperature (maximum phage titer at 37ºC and minimum at 4ºC); B, Comparative analysis of phage titer at various pH ranges (maximum titer obtained at 9 and minimum at 4); C, Comparative analysis of phage titer at various ranges of chloroform
4.6. One-Step Growth Curve
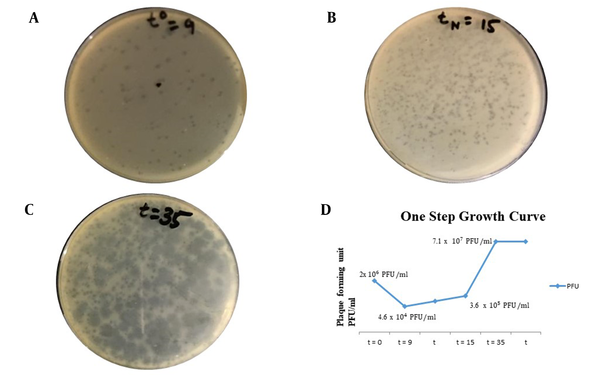
The results were obtained at an MOI of 0.01, as shown in Figure 2. The latent period of isolated bacteriophage was 24 minutes (which is 15 minutes for the first rise in titer after the adsorption period of 9 minutes). The burst size was determined as 190 phage particles per bacterial cell (190 PFU/cell). By adsorption assay, it was determined that after 9 minutes the bacteriophage was adsorbed to susceptible bacteria. Figure 4 depicts the graphical representation of the one-step growth curve.
A, One-step growth curve analysis (t0 referring to adsorption period = 9 minutes); B, tN = 15 minutes referring to time for first rise in phage titer after adsorption period (latent period = 9 + 15 = 24 minutes); C, t = 35 referring to time required to obtain maximum phage titer as no host remaining for reinfection and phage titer becoming constant (plateau period or burst time); D, Graphical presentation of one-step growth curve showing phage titer against K. pneumoniae (ie, host bacterium) at various time intervals
5. Discussion
The current study aimed to determine the prevalence of hvKp in clinical settings and isolate and characterize bacteriophage against this MDR hvKp as efficient naturally occurring alternative therapeutics. The hvKp is particularly associated with CAIs, and its incidence in clinical settings is increasing; nevertheless, the elevated drug resistance rates in hvKp are mainly associated with mobile genetic elements carrying resistant genes (8). Worldwide hvKp has been documented as the leading cause of numerous multisystem infections, including pneumonia, hepatic and nonhepatic abscesses, endophthalmitis, meningitis, skin and soft tissue infections, and necrotizing fasciitis (38). Consequently, the inevitable public health threat that emerged is the confluence of carbapenem resistance class (A and B) with the hypervirulent phenotype of K. pneumoniae and, more recently, the acquisition of mcr-1 for colistin resistance which is considered the last resort antibiotic against carbapenem-resistant pathogens (39).
In the current study, out of 50 samples, 22 samples (45%) were detected as K. pneumoniae. Overall, 11 isolates (50%) of K. pneumoniae were detected as hvKp; nevertheless, among 14 MDR K. pneumoniae isolates, 5 isolates (35%) were detected as MDR hvKp. All 5 MDR hvKp isolates (100%) were proven to be susceptible to our isolated bacteriophage. Among the hvKp isolates from various samples, maximum was detected in infected puss; however, only 1 hvKp was detected from tracheal aspirates. This unconventional association of hvKp with nonhepatic abscess is mainly due to the predominant expression of iron acquisition siderophores and polysaccharides (40). Furthermore, elevated rates of resistance were detected in hvKp isolates. However, previously the majority of hvKp were nonsusceptible to most of the antibiotics in comparison to their counterpart phenotype that largely possesses MDR and XDR isolates (8).
Aligned to the findings of the present study, recent studies, particularly from China, demonstrated the elevated prevalence of hvKp isolates, particularly resistant to carbapenems and colistin, detected in tertiary healthcare settings (41). Previously, two bacteriophages were isolated against hvKp, and both phages efficiently lysed hvKp that belonged to capsular serotype K57. However, the bacteriophages did not show any lytic activity against other capsular types indicating a limited host range (42). In line with the present study’s findings, previously isolated bacteriophage showed a short latent period with a large burst of lytic phage against hvKp (16).
High resistance levels were determined against every class of antibiotics, including more than 50% resistance to carbapenems (ie, doripenem 63% and 50% resistance rates determined against imipenem). Among cephalosporins, the maximum resistance rate was determined against ceftriaxone (81%) and cefotaxime (77%), followed by cefepime (77%) and ceftazidime (72%). Despite the limited data available for the resistance patterns of colistin, the current study demonstrated more than 22% colistin resistance which is 10 times higher than the previous study for colistin resistance (43). In this study, resistance ratios to imipenem, ciprofloxacin, and gentamycin were determined as 50%, 54%, and 54% as higher resistance ratios than the same resistance ratios of antibiotic classes recently determined in South Korea as 24%, 24%, and 3.1%, respectively (44). In contrast to the findings of present study, the lower resistance ratio of quinolones ie, ciprofloxacin and aminoglycosides is particularly due to their structural dissimilarities to cephalosporins.
The resistance patterns of this study are in great comparison to those of a recently conducted study in Pakistan, where maximum resistance was determined in K. pneumoniae isolates against ceftriaxone (71%). Additionally, nearly 40% resistance was determined against carbapenems, namely meropenem and imipenem. The minimum resistance was determined against colistin which was about 15%; however, 22% colistin resistance was determined in isolates collected in the present study (45). In comparison to the previous study conducted in Pakistan during 2010, this study reported 50% resistance to carbapenems which means a 50% MDR prevalence ratio, higher than previously noted (46). An increase in the overall burden of MDR prevalence ratio was also observed in countries, including India and China (47). The high resistance rates against carbapenems and high prevalence will lead to the usage of last resort beta-lactam monobactam, which in turn increases the resistance against monobactam (aztreonam). This statement can be proven by the recently determined resistant patterns against aztreonam which was the third-highest of the study after cephalosporins and carbapenems (45).
Hospital wastewater chosen for phage sample collection was inspired by the coevolution of bacteriophages and the associated host bacterium occurring in these environments (48). The coevolution of host and bacteriophages increases the exposure to different hosts, which in turn could increase the host range of bacteriophages present in such environments (49). Therefore, this study demonstrated that the sewage waste of hospitals could be a rich source for the isolation of bacteriophages with a broad host range. Such bacteriophages with a broad host range were previously used to treat MDR infections (50). Out of 22 isolates of K. pneumoniae, the isolated bacteriophage was active against 16 isolates (72%) of K. pneumoniae. Among 16 susceptible bacteria, 12 isolates were MDR K. pneumoniae. In contrast, previously isolated K. pneumoniae phages KP1513 and KP-34 showed a narrow host range (51). The most appropriate explanation of the broad host range of the isolated bacteriophage is based on the fact that K. pneumoniae phages are equipped with multiple depolymerasesthat play a critical role in the polyvalency of a particular bacteriophage (52).
Other features of the isolated bacteriophage included its adsorption period of 10 minutes and latent period of 25 minutes with a burst time of 55 minutes. This short latent period with a large burst size is a fundamental characteristic of in vivo phage therapy. The short latent period might allow the bacteriophage to clear the pathogen before the immune response mounted against the phage (53). The calculated burst size of the bacteriophage was estimated as 230 PFU/cell. The plaque size of the bacteriophage was estimated as 0.5 - 1 mm. The isolation of the bacteriophage with a broad host range requires early enrichment with multiple strains of bacteria. The same approach was adopted in this study to enrich bacteriophages with multiple bacterial isolates as previously hypothesized (49). Consequently, the isolated phages have shown a broad host range against MDR K. pneumoniae isolates with 77% activity against MDR isolates. Similar results were also obtained in a recent study on MDR K. pneumoniae bacteriophages with a 96% host range against carbapenem MDR K. pneumoniae (30). However, bacteriophages isolated from Italy and Australia showed narrow host ranges. The bacteriophages vB_Kpn_F48 (MG746602) from Italy and AmPh_EK29 (MN434092) from Australia were only active against strains of sequence type belonging to ST ST101 (16).
The isolated bacteriophages have shown stability at various ranges of temperature, pH, and chloroform. Similar results to the results of the present study have previously been obtained from bacteriophages that show stability to a wide pH range of 5 - 9 and retained residual activity at pH 3 (54). In this study, the estimated latent period was 24 minutes with a burst size of 140 phages per cell. However, the recent study showed that the Klebsiella bacteriophage had a latent period of 20 minutes which is in line with this study (latent period: 24 minutes); nevertheless, the burst size of that bacteriophage was 80 PFU/cell (6). The burst size was 5 times lower than the current study in which the burst size of the bacteriophage was calculated at 190 PFU/cell. One of the possible reasons for the small burst size is the morphology of bacteriophages; large phages usually have small burst sizes (6). The phage isolated in this study showed a broad host range with a high burst size suggesting that it might be used as an alternative therapeutic against MDR K. pneumoniae.
5.1. Conclusions
In conclusion, the emergence of MDR hvKp, particularly associated with HAIs and CAIs, demands alternative therapeutic approaches, such as phage therapy. The phage isolated in this study displayed efficient lytic potential with a broad host range. The stability in various environmental factors and short latent periods are the promising characteristics of the isolated phage. In the future, it is suggested to investigate the structural and molecular factors responsible for a broad host range and their potential use to develop engineered bacteriophages against MDR pathogens.
![A, Pink mucoid colonies of <i>K. pneumoniae</i> on MacConkey agar; B, Antibiotic susceptibility test results on Mueller-hinton agar plates by disk diffusion method (using antibiotic disks and Clinical and Laboratory Standards Institute guidelines [2020] for <i>K. pneumoniae</i>); C, String test showing the confirmed hypervirulent <i>Klebsiella pneumoniae</i> isolates; D, Comparative analysis of resistance against different antibiotics (maximum resistance against ceftriaxone in gray and minimum resistance against colistin in blue) A, Pink mucoid colonies of <i>K. pneumoniae</i> on MacConkey agar; B, Antibiotic susceptibility test results on Mueller-hinton agar plates by disk diffusion method (using antibiotic disks and Clinical and Laboratory Standards Institute guidelines [2020] for <i>K. pneumoniae</i>); C, String test showing the confirmed hypervirulent <i>Klebsiella pneumoniae</i> isolates; D, Comparative analysis of resistance against different antibiotics (maximum resistance against ceftriaxone in gray and minimum resistance against colistin in blue)](https://services.brieflands.com/cdn/serve/315dc/93527194d076a09b5f7edec454a96addc8ff711a/jjm-15-1-120027-i001-preview.png)