1. Background
Pseudomonas aeruginosa obligates aerobic Gram-negative bacillus and is the second most common cause of infective diseases in different hospitals’ wards including intensive care units. This organism becomes resistant to antimicrobial agents via various mechanisms, including altering the microorganism’s permeability against drugs, enzymatic resistance, efflux pump, receptor altering for drugs, and achieving a secondary metabolic pathway (1). Resistance to antibiotics in this bacterium occurs through mutation or further horizontal transfer through conjugation by plasmid and the transposons carrying antibiotic resistance genes (1, 2).
In this case, the plasmids and transposons carry antibiotic resistance genes and transfer them from one cell to another. In 1986, a DNA sequence with genes for resistance to different antibiotics was found. These regions have been identified at the other sites of various plasmids and are similar to transposons. Integrons are genetic units encompassing genes and transferring them while placed inside mobile factors, called gene cassettes (3). They have an integrase gene (int1), one member of the tyrosine recombinase family (4, 5). The integron structure consists of two fixed protected areas, called 3' and 5', located on both sides of a variable region (genetic cassette) containing one or several antibiotic resistance genes (6, 7). Gene cassettes usually contain one drug resistance gene and one protected site, providing the grounds for integrase identification through attachment and cutting processes.
Further, various studies have reported that integrons usually have more than one gene cassette, in which case these bacterial isolates have the multidrug resistance (MDR) pattern (4, 8). Gram-negative bacilli containing int1and its gene cassettes have aroused concerns among physicians and infection specialists regarding infection control. The resistance of this bacterium to a wide range of antibiotics, especially the common antibiotics used in hospitals, has complicated the detection of proper therapeutic methods and the use of infection control instruments (8, 9).
2. Objectives
This study aimed to detect the frequency of int1 and gene cassettes in the clinically isolates of P. aeruginosa from Babol, north of Iran, and investigate their correlations with antibiotic resistance patterns in hospitalized patients.
3. Methods
3.1. Clinical Samples
In this cross-sectional study, 30 clinical samples of P. aeruginosa were collected from patients admitted to Ayatollah Rohani Hospital, Babol, north of Iran, from 2016 to 2017. This project was a part of another project (10) (Code of Ethics: MUBABOL. REC.1394.162). The bacterial isolates were collected from the hospital laboratory. Pseudomonas aeruginosa was recognized using conventional biochemical and microbiological tests.
3.2. Antibiotic Susceptibility Test
Following the Clinical and Laboratory Standards Institute (CLSI document M100) guideline (11), antimicrobial susceptibility was determined by the Mueller-Hinton agar plates (Merck, Germany) using the standard disk diffusion (DD) method for antimicrobials under gentamicin (GM, 10 µg), cefepime (CPM, 30 µg), amikacin (AK, 30 µg), ciprofloxacin (CIP, 5 µg), imipenem (IMI 10 µg), cefotaxime(CTX, 30 µg), ampicillin (AP, 10µg), trimethoprim ( TM, 5 µg), and nitrofurantoin (NI, 300µg) (MAST Diagnostics, Merseyside, UK). Pseudomonas aeruginosa ATCC 27853 was used as a positive quality control (11-13).
3.3. Antibiotic Susceptibility Assay by Agar Dilution Method
After preparing stock solution according to the CLSI 2020 (11) standard for antibiotics, 1.5 × 108 CFU/mL of microbial suspensions were cultured in specimens on Muller-Hinton agar containing the concerned antibiotics (MAST Diagnostics, Merseyside, UK) and incubated at 37°C for 18 - 24 h. A plate containing medium without antimicrobial agents was considered a negative quality control, and P. aeruginosa ATCC 27853 strain was used as a positive quality control. The results were evaluated according to the table of the CLSI2020 standard (11-14).
3.4. DNA Extraction
The DNA extraction of all strains was performed using the PCR kit (Roche, Germany). The extracted DNAs were stored at -20°C for subsequent steps (12, 13, 15, 16).
3.5. Polymerase Chain Reaction Method (PCR)
Genomic DNA was extracted from P. aeruginosa colonies using the DNA purification kit (Roche, Germany). Table 1 presents the primer sequences. The total volume of the PCR reaction mixture was 60 µL encompassing 10 µL of extracted template DNA, 5.0 µL of 10x buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 50 pMole of each primer (Copenhagen, Denmark),1.5 U of Taq DNA polymerase (Amplicon Co., Denmark), and 21.8 µL ddH2O. Amplification was performed in a thermocycler (Corrbet, Australia) with a specific program (Table 1) and subjected to electrophoresis in 1.5% agarose gel. Consequently, the DNA standards of the PCR product were sequenced (Forster, USA). Standard strain ATCC1209 was used as a positive quality control and ATCC1053 as a negative quality control (15-18).
| Gene | Sequence Primer PCR Product | PCR Product | PCR Program |
|---|---|---|---|
| IntI | F: TCTCGGGTAACATCAAGG; R: AGGAGATCGGAAGACCTC | 243 bp | 5 min at 94°C; 35 cycles (1 min at 94°C, 1 min at 53°C, and 30 sec at 72°C); 5 min at 72°C |
| aadB | F: GGGCGCGTCATG GAG GAGTT; R: TATCGCGACCTGAAAGCGGC | 329 bp | 5 min at 94°C; 35 cycles (1 min at 94°C, 1 min at 65°C, and 30 sec at 72°C); 5min at 72°C |
| DfrA1 | F: GGAGTGCCAAAGGTGAACAGC; R: GAGGCGAAGTCTTGGGTAAAAAC | 367 bp | 5 min at 94°C; 35 cycles (1 min at 94°C, 1 min at 45°C, and 30 sec at 72°C); 5 min at 72°C |
| bla-OXA30 | F: ATTATCTACAGCAGCAGCGCCAGTGCATC; R: TTCGACCCCAAGTTTCCTGTAAGTGC | 716 bp | 5 min at 94°C; 35 cycles (1 min at 94°C, 1 min at 50°C, and 30 sec at 72°C); 5 min at 72°C |
3.6. Sequencing of Class 1 Integron Gene
The data of variable region of type 1 integron amplicons and its nucleotide sequences were sequenced and analyzed, respectively, by the National Center for Biotechnology Information (NCBI) (Retrieved from: http://www.ncbi.nlm.nih.gov/BLAST/).
3.7. Statistical Analysis
Paired-sample t test and Spearman’s correlation tests were used to analyze the collected data with SPSS software version 21.01. In this study, P < 0.05 was set as the significance level.
4. Results
4.1. Bacterial Isolation
During one year (2016 - 2017), 30 clinical isolates of P. aeruginosa were collected from patients admitted to Ayatollah Rohani Hospital (Babol, north of Iran).
4.2. Antibiotic Resistance Profile
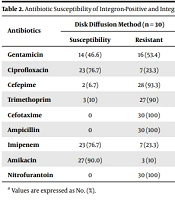
All samples were examined for resistance to nine antimicrobials using DD. The resistance rates to NI, AP, CTX, CPM, TM, GM, CIP, IMI, and AK were 100, 100, 100, 93.3, 9, 13.7, 10.3, and 3.4%, respectively (Table 2).
| Antibiotics | Disk Diffusion Method (n = 30) | Integron-Positive (n = 18) | Integron-Negative (n = 12) | P-Value | |||
|---|---|---|---|---|---|---|---|
| Susceptibility | Resistant | Susceptibility | Resistant | Susceptibility | Resistant | ||
| Gentamicin | 14 (46.6) | 16 (53.4) | 7 (38.9) | 11 (61.1) | 7 (58.3) | 5 (41.7) | 0.296 |
| Ciprofloxacin | 23 (76.7) | 7 (23.3) | 13 (72.2) | 5 (27.8) | 10 (83.3) | 2 (16.7) | 0.481 |
| Cefepime | 2 (6.7) | 28 (93.3) | 1 (5.6) | 17 (94.4) | 1 (8.3) | 11 (91.7) | 0.795 |
| Trimethoprim | 3 (10) | 27 (90) | 0 | 18 (100) | 3 (25.0) | 9 (75.0) | 0.025* |
| Cefotaxime | 0 | 30 (100) | 0 | 18 (100) | 0 | 12 (100) | 0.00* |
| Ampicillin | 0 | 30 (100) | 0 | 18 (100) | 0 | 12 (100) | 0.00* |
| Imipenem | 23 (76.7) | 7 (23.3) | 14 (77.8) | 4 (22.2) | 9 (75.0) | 3 (25.0) | 0.860 |
| Amikacin | 27 (90.0) | 3 (10) | 14 (77.8) | 4 (22.2) | 11 (91.7) | 1 (8.3) | 0.804 |
| Nitrofurantoin | 0 | 30 (100) | 0 | 18 (100) | 0 | 12 (100) | 0.00* |
a Values are expressed as No. (%).
The results of the agar dilution method rates for NI, AP, CTX, TM, GM, CPM, CIP, IMI, and Ak were 100, 96.7, 96.7, 90, 63.3, 56.7, 40, 40, and 3.3%, respectively (Table 3). The multidrug resistance phenotype of P. aeruginosa clinical isolates for intI and gene cassettes of aadB, dfrA1, and bla-OXA30 are presented in Table 4.
| Antibiotic | Agar Dilution Method (n = 30) | Integron-Positive (n = 18) | Integron-Negative (n = 12) | P-Value | |||
|---|---|---|---|---|---|---|---|
| Susceptibility | Resistant | Susceptibility | Resistant | Susceptibility | Resistant | ||
| Gentamicin | 11 (36.7) | 19 (63.3) | 3 (16.7) | 15 (83.3) | 8 (66.6) | 4 (33.4) | 0.005 * |
| Ciprofloxacin | 18 (60) | 12 (40) | 8 (44.4) | 10 (55.6) | 10 (83.3) | 2 (16.7) | 0.033* |
| Cefepime | 13 (43.3) | 17 (65.7) | 3 (16.7) | 15 (83.3) | 10 (83.3) | 2 (16.7) | 0.000* |
| Trimethoprim | 3 (10) | 27 (90) | 0 | 18 (100) | 3 (25) | 9 (75) | 0.025* |
| Cefotaxime | 1 (3.3) | 29 (96.7) | 0 | 18 (100) | 1 (8.3) | 11 (91.7) | 0.213 |
| Ampicillin | 1 (3.3) | 29 (96.7) | 0 | 18 (100) | 1 (8.3) | 11 (91.7) | 0.213 |
| Imipenem | 18 (60) | 12 (40) | 11 (61.1) | 7 (38.9) | 7 (58.3) | 5 (41.7) | 0.879 |
| Amikacin | 29 (96.7) | 1 (3.3) | 17 (94.4) | 1 (5.6) | 12 (100) | 0 | 0.406 |
| Nitrofurantoin | 0 | 30 (100) | 0 | 18 (100) | 0 | 12 (100) | 0.000* |
a Values are expressed as No. (%).
| Antibiotics Resistant Pattern | Number of Isolates | IntI | aadB | DfrA1 | bla-OXA30 |
|---|---|---|---|---|---|
| CTX/SXT/AM/IPM/FM/GM/CP/FEP | 1 | 1 | - | 1 | - |
| CTX/SXT/AM/AN/FM/GM/FEP | 2 | 2 | 2 | 1 | 1 |
| CTX/SXT/AM/IPM/FM/GM/FEP | 2 | 2 | 2 | 2 | - |
| CTX/SXT/AM/IPM/FM/GM/CP | 1 | 1 | - | 1 | - |
| CTX/SXT/AM/FM/GM/CP/FEP | 1 | - | - | - | - |
| CTX/SXT/AM/FM/GM/FEP | 8 | 5 | 3 | 2 | 2 |
| CTX/SXT/AM/AN/FM/FEP | 1 | 1 | 1 | 1 | - |
| CTX/SXT/AM/FM/CP/FEP | 3 | 3 | - | 1 | - |
| CTX/SXT/AM/IPM/FM/FEP | 3 | 1 | 1 | 1 | 1 |
| CTX/SXT/AM/FM/FEP | 5 | 2 | 2 | 2 | 2 |
| CTX/AM/FM/CP/FEP | 1 | - | - | - | - |
| CTX/AM/FM/GM | 1 | - | - | - | - |
| CTX/AM/FM/FEP | 1 | - | - | - | - |
4.3. Gene Cassettes
60% (n : 18) carried intI gene among 30 P. aeruginosa isolates. The prevalence of aadB, dfrA1, and bla-OXA30 genes were 11 (61%), 12 (66%), 6 (33%), and 0 (0%), respectively.
4.4. Nucleotide Sequence Accession Number
The positive aadB and dfrA1strains were sequenced and established in the Gene Bank database as MH708573 and MH708574, respectively.
5. Discussion
Pseudomonas aeruginosa is an important opportunistic nosocomial bacterium with high resistance to most common antibiotics. For this reason, treating infections induced by this bacterium is challenging. The innate and acquired resistance to antimicrobial agents is involved in the mortality rate of patients suffering from infections induced by this bacterium (19). Integrons have been known as the primary mechanism to detect the gene of resistance to antibiotics in Gram-negative bacteria (20). The integrase gene sets the ground for resistance to common antibiotics, including ampicillin, gentamicin, trimethoprim, and cefotaxime (5). The findings of this study indicated that 18 isolates (60%) had the int1 gene. Further, out of the samples with the int1 gene, 61, 62, and 33% had the gene cassettes of aadB, dfrA1, and bla-OXA30.
The correlation between the presence of the intI gene and inducing resistance to each antibiotic was examined with SPSS software version 21.0 using the chi-square test. In the DD method, there is a significant correlation between the presence of intI gene and resistance to trimethoprim, cefotaxime, ampicillin, and nitrofurantoin. In the agar dilution method, there is a significant correlation between the presence of intI gene and resistance to gentamicin, ciprofloxacin, cefepime, trimethoprim, and nitrofurantoin separately (Table 2,3). In Brazil, Fonseca et al. (21) documented 45.5% of P. aeruginosa strains had the int1 gene, and out of 29 isolates, 66% and 52% samples had aacA and bla-OXA30 genes. In another study, the intI gene frequency was 57.1%, and there was also a significant relationship between the gene cassettes in int1 and antibiotic resistance (22). In another study by Nikokar et al. (23) on antibiotic resistance of the Int1 gene in P. aeruginosa, 47% of the strains were resistant to the antibiotic. However, most of the strains were insensitive to imipenem. The PCR results indicated that 45.3% of the isolates had the intI gene and that 69% of the strains were antibiotic-resistant. All studies suggest a significant relationship between the presence of int1 and gene cassettes with antibiotic resistance (24).
5.1. Conclusions
The high resistance of P. aeruginosa isolates is due to the presence of int1 and its gene cassettes. Considering their high resistance to cefotaxime, gentamicin, ampicillin, and imipenem in hospitals, selecting appropriate drugs or generally changing the treatment course for patients is possible to prevent the spread of resistance inducing genes and the development of nosocomial infections.