1. Background
Diphtheria is still a global health concern, especially in low- and middle-income countries. The statistics reported by the World Health Organization (WHO) indicates that countries with the highest frequency of diphtheria cases during the last five years (2015 - 2019) were India, Ethiopia, Nigeria, Madagascar, Indonesia, Yemen, Pakistan, Venezuela, and Nepal. In this regard, Indonesia is consistently among the five countries with the highest frequency of diphtheria cases each year (1). Data from the Ministry of Health of the Republic of Indonesia shows that several regions such as East Java, West Java, Jakarta, and Banten encompassed the largest number of diphtheria cases (2). Diphtheria is a serious health problem because of its high case fatality rate (CFR) (from about 10% to > 50% in severe cases) (3). Moreover, this disease is proved to spread across regions, islands, and even countries (4).
Diphtheria is caused by Corynebacterium diphtheriae and two closely related species, called C. ulcerans and C. pseudotuberculosis (5). Corynebacterium diphtheriae is the leading cause of diphtheria and is widely isolated in developing countries. Corynebacterium ulcerans is more often isolated in developed countries with high immunization coverage. Meanwhile, C. pseudotuberculosis has rarely caused diphtheria in humans in developed and developing countries (6). The potential of these bacteria to produce diphtheria toxin makes them different from the other species of the genus Corynebacterium. Diphtheria toxin is a major virulence factor for diphtheria-causing bacteria and is associated with the clinical features of diphtheria disease. However, not all strains of diphtheria-causing bacteria can produce toxins (toxigenic). In nontoxigenic strains, no tox gene encoding toxin synthesis has been found. Nontoxigenic strains do not cause diphtheria diseases; however, their presence needs to be continuously monitored because they can cause other severe diseases such as endocarditis or bacteremia. Nontoxigenic strains can also change to toxigenic and cause diphtheria when the bacterial chromosome is inserted by a typical Corynephage carrying the tox gene (7, 8).
Although the diagnosis and treatment of diphtheria are based on clinical manifestation, some cases are mild and atypical. Laboratory tests are needed to rule out a differential diagnosis (9, 10). The fast and accurate laboratory tests help prevent the spread of diphtheria-causing bacteria and provide case management as early as possible. However, the gold standard for the laboratory examination of diphtheria using conventional methods has several limitations. The conventional methods were completed in 3 - 5 days, and the results were affected by sample transportation and history of antibiotic administration. This is a major problem for countries such as Indonesia. In this case, PCR can be an alternative, which is fast and relatively less affected by sample delivery problems and history of antibiotic use (10). Diphtheria laboratory examination using the PCR method has long been introduced as a screening method to examine toxigenicity. Some PCR assays focus on detecting toxigenic strains, while others include the identification of nontoxigenic strains (11, 12). However, PCR usually cannot be used to distinguish between toxigenic and non-toxigenic tox gene bearing (NTTB) strains. We previously succeeded in developing PCR to predict the NTTB strains; however, it is still limited to two types, namely base-25 deletion or base-55 deletion NTTB (13).
2. Objectives
The study aimed to develop and implement a multiplex real-time PCR to identify three species of diphtheria-causing bacteria and screen their toxigenicity quickly and accurately using the dtxR and tox genes as the target.
3. Methods
3.1. PCR Primers and Probes
The multiplex real-time PCR in this study used two target genes: the dtxR gene for species identification and the tox gene for bacterial toxigenicity screening (Table 1). Only two pairs of the PCR primers were used according to the target genes. Meanwhile, the used probe consisted of one probe for the tox gene and three other probes for the dtxR gene of C. diphtheriae, C. ulcerans, and C. pseudotuberculosis with a competitive system. Accordingly, the probe was designed to distinguish three species of diphtheria-causing bacteria regarding the differentiation of dtxR gene sequences. The PCR primers and probes were designed semi-manually by aligning the sequences of tox and dtxR genes of three species of diphtheria-causing bacteria. Sequence alignment was carried out using BioEdit on the variations of dtxR and tox genes of three diphtheria-causing bacteria. A pair of the PCR primers and a probe targeting the tox gene were designed in the conserved and specific regions for all three species and the pairs of PCR primers targeting the dtxR gene. Some parameters, including G-C content, Tm, run, and repeat, were obtained with PerlPrimer software. The specificity of the PCR primer and probe was analyzed with online Primer-BLAST. The delta G (free energy) value of the self-dimer or cross-dimer at the 3′ end was not < -7 kcal/mol (14).
3.2. Samples
The research sample encompassed seven reference strains of Corynebacterium spp, a synthetic DNA, 30 archived isolates, and 924 clinical specimens. The reference strains consisted of three isolates of toxigenic C. diphtheriae (ATCC 13812, NCTC 10648, and NCTC 3984), one isolate of non-toxigenic C. diphtheriae (NCTC 10356), one isolate of non-toxigenic C. ulcerans (NCTC 12077), one isolate of C. striatum (NCTC 764), and one isolate of C. minutissimum (ATCC 23348). A synthetic double-stranded DNA (gBlock, Integrated DNA Technologies, Inc.) identical to the dtxR gene of C. pseudotuberculosis 316 was used to substitute the C. pseudotuberculosis isolate not available in this study.
The archive isolates were as follows: 10 isolates of toxigenic C. diphtheriae, one isolate of non-toxigenic C. diphtheriae, and 19 isolates of non-diphtheria-causing bacteria (namely Neisseria meningitidis, N. gonorrhoeae, Streptococcus pneumoniae, S. agalactiae, Staphylococcus aureus, S. epidermidis, Klebsiella pneumoniae, Legionella pneumophila, Enterobacter sakazakii, Pseudomonas aeruginosa, Mycobacterium tuberculosis, Salmonella typhimurium, Haemophilus influenzae b, Shigella flexneri, Aeromonas hydrophila, Vibrio cholerae, Escherichia coli, Clostridium tetani, and Candida albicans). The C. diphtheriae and C. ulcerans isolates and synthetic DNA were used as a positive control, while the others were used as a negative control in the PCR optimization method. The clinical specimen in this study were throat swabs sent to the laboratory for diphtheria examination. In this study, 924 throat swabs were collected from 311 diphtheria clinical cases and 613 close contacts. The clinical cases were diagnosed by a physician when the signs and symptoms matched the diphtheria criteria described in the National Diphtheria Surveillance Guideline.
3.3. Conventional Methods
The reference strains and the archived isolates were revived and re-identified by microscopic and biochemical examination and Elek tests. On the other hand, the clinical specimens were identified using conventional methods according to the WHO guidelines for diphtheria laboratory (15). The diphtheria-causing bacteria were identified using API Coryne® (bioMérieux), while the bacterial toxigenicity was determined using modified Elek tests (16). After the clinical specimens were examined using the conventional methods, the samples were transferred to 0.5 mL Aquadest in the microtube for the DNA extraction.
3.4. Multiplex Real-time PCR
DNA was extracted using the commercial QIAamp DNA Mini Kit (Qiagen) (17). The DNA in 50 uL molecular water was stored at -20°C for the DNA template in the PCR examination. The multiplex real-time PCR process was run in the CFX 96 thermal cycler (Biorad). The PCR mix consisted of 10 uL SensiFASTTM Probe No-ROX (Bioline), 0.5 uL each PCR primer, 0.5 uL each probe, 1 uL ddH2O, and 5 uL DNA template. The PCR conditions followed an initial denaturation phase at 95°C for 3 min, with 35 cycles of a denaturation phase at 95°C for 10 s and annealing and extension phases at 60°C for 30 s. The PCR results were interpreted according to the sigmoid curve and Ct value of each fluorophore. The Cy5 fluorophore was used as a marker for C. diphtheriae, while Hex and FAM fluorophores were used as markers for C. pseudotuberculosis and C. ulcerans, respectively. For bacterial toxigenicity markers, a Texas red fluorophore was used. When a sigmoid curve appeared with a Ct value < 36 on the Cy5 and Texas red fluorophores, it was concluded as toxigenic C. diphtheriae. When only Cy5 appeared, it was concluded as nontoxigenic C. diphtheriae. The same applied to the Hex fluorophore for C. pseudotuberculosis and the FAM fluorophore for C. ulcerans. If only the Texas red fluorophore is present, the test should be repeated.
3.5. Established Conventional PCR and Data Analysis
The established conventional PCR assay was carried out to ensure the accuracy of the multiplex real-time PCR assay. The procedure of the conventional PCR assay was according to the protocol described in the previous study (13). The multiplex real-time PCR results were considered accurate when the results were similar to the established conventional PCR results, even though the conventional methods showed different results.
4. Results
4.1. PCR Primers and Probes
The developed multiplex real-time PCR in this study used two pairs of PCR primers and four Taqman probes (Table 1). Primer-dimer binding risks were decreased by minimizing the number of the PCR primers. The sequences analysis showed acceptable criteria for the primer and probe, including the presence of conserved and specific sequences with 40 - 60% GC content (Data is not presented here).
| PCR; Primers & Probes | Sequence (5′ - 3′) | Gene | Target |
|---|---|---|---|
| DIP_UL_PSE_ F | CCTACAGTTAGCCAAACMGTTGC | dtxR | Corynebacterium diphtheriae, C. ulcerans, C. pseudotuberculosis |
| DIP_UL_PSE_R | CGGCAGGCTTCATCGTGMAC | dtxR | C. diphtheriae, C. ulcerans, C. pseudotuberculosis |
| DIPHTHER | Cy5-ACTTGTCGTTGTCGCcTCaGACC-BHQ2 | dtxR | C. diphtheriae |
| ULCER | FAM-TAGTCGCaTCcGACCGCAGC-BHQ1 | dtxR | C. ulcerans |
| PSEUDOT | Hex-GTAGTTGCgTCtGACCGTAGTCTTCAA-BHQ1 | dtxR | C. pseudotuberculosis |
| TOX_B_ F | CAGTTGGAACACTGTTGAAGATTCGAT | tox | C. diphtheriae, C. ulcerans, C. pseudotuberculosis (toxigenic) |
| TOX_B_R | CGACCATTTACGGAAATATGAGTCTTGGAC | tox | C. diphtheriae, C. ulcerans, C. pseudotuberculosis (toxigenic) |
| TOX_B | Texas Red-AACGTCCAGCTTTCCAGGAATAGTCG-BHQ2 | tox | C. diphtheriae, C. ulcerans, C. pseudotuberculosis (toxigenic) |
4.2. Conventional Methods
The re-identification of isolates showed that the results matched the data of the isolates (Table 2). The identification of the clinical specimens using conventional methods showed that the diphtheria-causing bacteria (C. diphtheriae) were isolated from 79 (25.4%) out of 311 and three (0.5%) out of 613 clinical samples isolated from diphtheria cases and close contacts, respectively. Eighty of the 82 isolates were identified as toxigenic using the modified Elek test; however, two isolates were non-toxigenic.
| Variables | Multiplex Real-time PCR | Established Conventional PCR | Conventional Methods |
|---|---|---|---|
| Reference isolates/synthetic DNA | |||
| Toxigenic Corynebacterium diphtheriae | 3 | 3 | 3 |
| Non-toxigenic C. diphtheriae | 1 | 1 | 1 |
| C. ulcerans | 1 | 1 | 1 |
| C. pseudotuberculosisa | 1 | 1 | NA |
| Negatif for diphtheria-causing bacteria | 2 | 2 | 2 |
| Total | 8 | 8 | 7 |
| Archived isolates | |||
| Toxigenic C. diphtheriae | 10 | 10 | 10 |
| Non-toxigenic C. diphtheriae | 1 | 1 | 1 |
| C. ulcerans/C. pseudotuberculosis | 0 | 0 | 0 |
| Negatif for diphtheria-causing bacteria | 19 | 19 | 19 |
| Total | 30 | 30 | 30 |
| Clinical samples | |||
| Toxigenic C. diphtheriae | 130 | 130 | 70 |
| Non-toxigenic C. diphtheriae | 2 | 2 | 2 |
| C.ulcerans/C.pseudotuberculosis | 0 | 0 | 0 |
| Negatif for diphtheria-causing bacteria | 792 | 792 | 852 |
| Total | 924 | 924 | 924 |
Abbreviation: NA, not applicable.
a Synthetic DNA.
4.3. PCR Assay
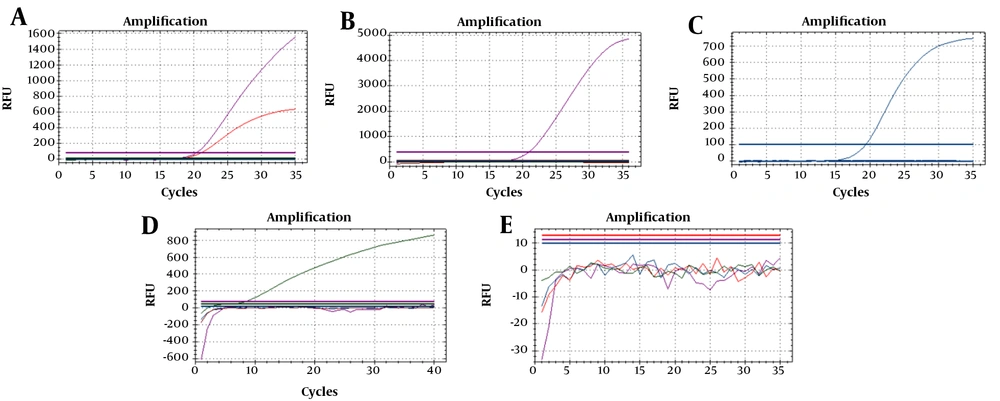
Seven reference strains and one synthetic DNA were used to optimize the PCR reaction (Figure 1). For three isolates of toxigenic C. diphtheriae (ATCC 13812, NCTC 10648, and NCTC 3984), the sigmoid curve for both fluorophores (Cy5 dan Texas red) appeared. In contrast, for one isolate of non-toxigenic C. diphtheriae (NCTC 10356), one isolate of non-toxigenic C. ulcerans (NCTC 12077), and one DNA synthetic of C. pseudotuberculosis, there was only one sigmoid curve of fluorophore (Cy5 for C. diphtheriae, FAM for C. ulcerans, and Hex for C. pseudotuberculosis). On the other hand, no sigmoid curve of fluorophore appeared for the two isolates of non-diphtheria-causing bacteria (NCTC 764 and ATCC 23348). Thirty archived isolates were accurately identified using a multiplex real-time PCR as there were 10 isolates toxigenic C. diphtheriae, one isolate non-toxigenic C. diphtheriae, and 19 isolates negative for diphtheria-causing bacteria (Table 2). The results also showed that 130 toxigenic C. diphtheriae and two non-toxigenic C. diphtherae were identified in 120 (38.6%) out of 311 clinical specimens from cases and 12 (2%) out of 613 clinical specimens from close contacts. No C. ulcerans and C. pseudotuberculosis were found in the clinical specimen in this study. The findings were similar to the established PCR results.
5. Discussion
This study developed a multiplex real-time PCR to identify diphtheria-causing bacteria, similar to previous studies by de Zoysa et al. and Badell et al. (18, 19). However, the dtxR gene was used in this study, while the aforementioned researchers used the rpoB gene. Previously, we showed that the dtxR gene had some advantages over 16S rRNA and pld genes in identifying C. ulcerans and C. pseudotuberculosis (20). Moreover, we detected and identified three species of diphtheria-causing bacteria, while they only identified two (C. diphtheriae and C. ulcerans/C. pseudotuberculosis).
Furthermore, we used a pair of the PCR primers to identify three species; however, they used more pairs. Using only a pair of PCR primers would decrease the risk of primer-dimer binding and reduce costs imposed by the PCR primer synthesis. We used the tox gene as a target to identify toxigenicity, similar to their study. Unlike our previous study (13), we did not identify the NTTB strains to reduce the cost in this study because of the high cost of the PCR probe. Furthermore, previous studies documented that NTTB strains were never observed in diphtheria samples in Indonesia (4, 10, 13). The NTTB strain originated in Indonesia was isolated from the non-diphtheria sample (21).
The sensitivity of multiplex real-time PCR developed in this study was quite acceptable. Regarding the limit of detection (LOD), the sensitivity was predicted by a 10-fold dilution of 0.5 Mc Farland bacteria cells up to 10-6 (approximately 30 CFU/200 uL bacteria cell or 30 CFU/50 uL DNA template). This means this method can detect target bacteria < 10 CFU/reaction. The sensitivity of PCR is also predicted based on its performance in identifying target bacteria in the clinical specimen. In this case, PCR was approximately 1.5 - 4 folds more sensitive than the conventional methods (38.6%: 25.4% for cases and 2%: 0.5% for close contacts). Previous studies showed that PCR was approximately two folds more sensitive than conventional PCR (11, 13). On the other hand, the efficiency of PCR in this study was in an acceptable range (90 - 110%) for four targets (data not shown). The PCR efficiency was estimated by E = 10[-1/slope] – 1, as described previously (22).
The specificity of the PCR developed in this study was also quite acceptable. The PCR could correctly identify C. diphtheriae, C. ulcerans, and C. pseudotuberculosis for reference strains and synthetic DNA. Unfortunately, we found no bacteria for the archived isolates or the clinical specimens using conventional methods and PCR. The differentiation of the results between PCR and conventional methods for the clinical specimens was not predicted due to specificity weakness or mismatching. It is supported by the similarity of results between multiplex real-time PCR and established PCR assay using different PCR primers (13). Similar to a previous study (13), the identification of diphtheria-causing bacteria in the clinical samples indicated that the causative agents of diphtheria in Indonesia were dominated by toxigenic C. diphtheriae. This is different from the findings for developed countries where non-toxigenic C. diphtheriae and C. ulcerans are widely isolated (23, 24). However, we still believe that C. ulcerans and C. pseudotuberculosis can cause health problems, and this issue needs to be addressed in the future in Indonesia.
5.1. Conclusions
The developed multiplex real-time PCR in this study can identify three species of diphtheria-causing bacteria and screen their toxigenicity fastly and accurately. However, in this study, no diphtheria-causing bacteria other than C. diphtheriae was found in the clinical samples using PCR or conventional methods. PCR is more sensitive than conventional methods and can be used as an additional test in diphtheria laboratories.